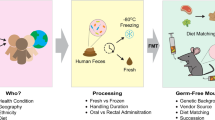
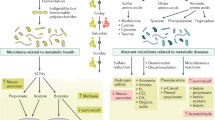
Abstract
Research into the gut microbiota and its role in health and disease has expanded rapidly in the past two decades. However, much of the early focus has been on cataloguing the microorganisms present, identifying correlations between microbial species and disease and using preclinical animal models to understand phenotypes. Now efforts are under way to provide functional insights into the gut microbiota and its mechanisms of action, improve understanding of the role of the microbiota beyond the gut and advance the development of microbiota-based therapeutics so that the microbiome can be harnessed in the clinic. In this Viewpoint article, we asked a selection of scientists and clinicians working in the gut microbiome field for their opinions on the major advances in and the challenges and solutions for translating gut microbiome research to the clinic, and where they expect progress to be made in the future.
The contributors
Susan V. Lynch is a professor in the Department of Medicine at the University of California, San Francisco and Director of the Benioff Center for Microbiome Medicine, United States. Her research focuses on the gut microbiome and chronic inflammatory disease, with specific emphasis on childhood allergy and asthma, and adult inflammatory bowel disease.
Siew C. Ng is a professor in the Department of Medicine and Therapeutics, Associate Director of the Centre for Gut Microbiota Research, Director of the Microbiota Innovation Centre (MagIC) and Assistant Dean for Development at the Chinese University of Hong Kong, Hong Kong. She is an Associate Editor of Gut. Her interest focuses on studies of the human gut microbiota in health and diseases, such as inflammatory bowel disease, cancer and obesity, and the increasing role of the virome and fungi in faecal microbiota transplantation.
Fergus Shanahan is Emeritus Professor of Medicine at University College Cork, National University of Ireland, and Foundation Director of APC Microbiome Ireland, a research centre funded by Science Foundation Ireland since 2003, which investigates host–microorganism interactions in the gut. His interests include most things that affect the experience of illness.
Herbert Tilg is Head of the Department of Gastroenterology, Hepatology, Endocrinology and Metabolism at Medical University Innsbruck, Austria. His main research interest is inflammation, innate immunity and interaction with the gut microbiome in gastrointestinal and liver disorders. He is an Associate Editor of Gut and Chairman of the Scientific Committee, United European Gastroenterology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Donia, M. S. & Fischbach, M. A. Small molecules from the human microbiota. Science 349, 1254766 (2015).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
Fujimura, K. E. et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191 (2016).
Levan, S. R. et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. https://doi.org/10.1038/s41564-019-0498-2 (2019).
Yu, T. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e16 (2017).
Norman, J. M. et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 (2015).
Zuo, T. et al. Gut mucosal virome alterations in ulcerative colitis. Gut. 68, 1169–1179 (2019).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Vangay, P. et al. US immigration westernizes the human gut microbiome. Cell 175, 962–972 e910 (2018).
Chassaing, B. et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 (2015).
Paramsothy, S. et al. Faecal microbiota transplantation for inflammatory bowel disease: a systematic review and meta-analysis. J. Crohns. Colitis. 11, 1180–1199 (2017).
Xu, D. et al. Efficacy of fecal microbiota transplantation in irritable bowel syndrome: a systematic review and meta-analysis. Am. J. Gastroenterol. 114, 1043–1050 (2019).
Schnadower, D. et al. Lactobacillus rhamnosus GG versus placebo for acute gastroenteritis in children. N. Engl. J. Med. 379, 2002–2014 (2018).
Freedman, S. B. et al. Multicenter trial of a combination probiotic for children with gastroenteritis. N. Engl. J. Med. 379, 2015–2026 (2018).
Ooijevaar, R. E., Terveer, E. M., Verspaget, H. W., Kuijper, E. J. & Keller, J. J. Clinical application and potential of fecal microbiota transplantation. Annu. Rev. Med. 70, 335–351 (2019).
Shanahan, F. The gut microbiota – a clinical perspective on lessons learned. Nat. Rev. Gastroenterol. Hepatol. 9, 609–614 (2012).
Shanahan, F., van Sinderen, D., O’Toole, P. W. & Stanton, C. Feeding the microbiota: transducer of nutrient signals for the host. Gut. 66, 1709–1717 (2017).
Murphy, C. L., O’Toole, P. W. & Shanahan, F. The gut microbiota in causation, detection, and treatment of cancer. Am. J. Gastroenterol. 114, 1036–1042 (2019).
Nadim, M. K. et al. Management of the critically ill patient with cirrhosis: A multidisciplinary perspective. J. Hepatol. 64, 717–735 (2016).
Backhed, F. et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl Acad. Sci. USA 101, 15718–15723 (2004).
Loomba, R. et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 25, 1054–1062 e1055 (2017).
Hoyles, L. et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 24, 1070–1080 (2018).
Grander, C. et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. 67, 891–901 (2018).
Puri, P. et al. The circulating microbiome signature and inferred functional metagenomics in alcoholic hepatitis. Hepatology 67, 1284–1302 (2018).
Hendrikx, T. et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 68, 1504–1515 (2019).
Kummen, M. et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut. 66, 611–619 (2017).
Nakamoto, N. et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol. 4, 492–503 (2019).
Manfredo Vieira, S. et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359, 1156–1161 (2018).
Qin, N. et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 (2014).
Bajaj, J. S. et al. Long-term outcomes of fecal microbiota transplantation in patients with cirrhosis. Gastroenterology 156, 1921–1923 e1923 (2019).
Bullman, S. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017).
Suez, J. et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423.e16 (2018).
Zmora, N. et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174, 1388–1405.e21 (2018).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Palleja, A. et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 3, 1255–1265 (2018).
Browne, H. P. et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546 (2016).
Goodman, A. L. et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl Acad. Sci. USA 108, 6252–6257 (2011).
Park, D. I. et al. Asian Organization for Crohn’s and Colitis and Asia Pacific Association of Gastroenterology consensus on tuberculosis infection in patients with inflammatory bowel disease receiving anti-tumor necrosis factor treatment. Part 2: management. Intest. Res. 16, 17–25 (2018).
Deschasaux, M. et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med. 24, 1526–1531 (2018).
Serrano, M. G. et al. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 25, 1001–1011 (2019).
Shan, Y., Segre, J. A. & Chang, E. B. Responsible stewardship for communicating microbiome research to the press and public. Nat. Med. 25, 872–874 (2019).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662,e20 (2019).
Suez, J., Zmora, N., Segal, E. & Elinav, E. The pros, cons, and many unknowns of probiotics. Nat. Med. 25, 716–729 (2019).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Duvallet, C., Gibbons, S. M., Gurry, T., Irizarry, R. A. & Alm, E. J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 8, 1784 (2017).
Sonnenburg, E. D. & Sonnenburg, J. L. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 17, 383–390 (2019).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Zuo, T. et al. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat. Commun. 9, 3663 (2018).
Zuo, T. et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 67, 634–643 (2018).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Zhou, W. et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 569, 663–671 (2019).
Lloyd-Price, J., Abu-Ali, G. & Huttenhower, C. The healthy human microbiome. Genome. Med. 8, 51 (2016).
Robinson, T. P. et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 110, 377–380 (2016).
Acknowledgements
S.V.L. is funded by awards from the US National Institutes of Health (P0515267, AI113916, AI133765, DH082147, DA040532), the Crohn’s and Colitis Foundation of America (598710) and the Rainin Foundation. S.C.N. acknowledges Z. Tao and W. Tang for their scientific input. F.S. is funded in part by Science Foundation Ireland (centre grant APC SFI/12RC/2273).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
S.V.L. cofounded Siolta Therapeutics and is a board member and consultant for this company. She also served on the scientific advisory board of Bloom Science. F.S. is a cofounder and shareholder of Alimentary Health, Tucana Health and Atlantia Food Clinical Trials. These facts neither influenced nor constrained the contents of his responses here. S.C.N. and H.T. declare no competing interests.
Rights and permissions
About this article
Cite this article
Lynch, S.V., Ng, S.C., Shanahan, F. et al. Translating the gut microbiome: ready for the clinic?. Nat Rev Gastroenterol Hepatol 16, 656–661 (2019). https://doi.org/10.1038/s41575-019-0204-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-019-0204-0
This article is cited by
-
Multi-factorial examination of amplicon sequencing workflows from sample preparation to bioinformatic analysis
BMC Microbiology (2023)
-
The microbiome–gut–brain axis in Parkinson disease — from basic research to the clinic
Nature Reviews Neurology (2022)
-
Multimodal interactions of drugs, natural compounds and pollutants with the gut microbiota
Nature Reviews Microbiology (2022)
-
Small phenolic and indolic gut-dependent molecules in the primate central nervous system: levels vs. bioactivity
Metabolomics (2022)
-
Gut microbiota alteration in hepatobiliary diseases: cause-and-effect relationship
Hepatology International (2021)