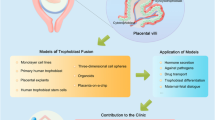
Abstract
The importance of the placenta in supporting mammalian development has long been recognized, but our knowledge of the molecular, genetic and epigenetic requirements that underpin normal placentation has remained remarkably under-appreciated. Both the in vivo mouse model and in vitro-derived murine trophoblast stem cells have been invaluable research tools for gaining insights into these aspects of placental development and function, with recent studies starting to reshape our view of how a unique epigenetic environment contributes to trophoblast differentiation and placenta formation. These advances, together with recent successes in deriving human trophoblast stem cells, open up new and exciting prospects in basic and clinical settings that will help deepen our understanding of placental development and associated disorders of pregnancy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Brosens, I., Pijnenborg, R., Vercruysse, L. & Romero, R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 204, 193–201 (2011).
Pijnenborg, R., Robertson, W. B., Brosens, I. & Dixon, G. Trophoblast invasion and the establishment of haemochorial placentation in man and laboratory animals. Placenta 2, 71–91 (1981).
Napso, T., Yong, H. E. J., Lopez-Tello, J. & Sferruzzi-Perri, A. N. The role of placental hormones in mediating maternal adaptations to support pregnancy and lactation. Front. Physiol. 9, 1091 (2018).
PrabhuDas, M. et al. Immune mechanisms at the maternal-fetal interface: perspectives and challenges. Nat. Immunol. 16, 328–334 (2015).
Moffett, A. & Loke, C. Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 6, 584–594 (2006).
Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A. & Rossant, J. Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 (1998). This study describes the ground-breaking derivation of mouse trophoblast stem cells, thereby providing a fundamental research tool that has substantially advanced our understanding of the mechanistic principles underlying trophoblast development.
Gamage, T. K., Chamley, L. W. & James, J. L. Stem cell insights into human trophoblast lineage differentiation. Hum. Reprod. Update 23, 77–103 (2016).
Cambuli, F. et al. Epigenetic memory of the first cell fate decision prevents complete ES cell reprogramming into trophoblast. Nat. Commun. 5, 5538 (2014).
Malassine, A., Frendo, J. L. & Evain-Brion, D. A comparison of placental development and endocrine functions between the human and mouse model. Hum. Reprod. Update 9, 531–539 (2003).
Okae, H. et al. Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e56 (2018). More than 20 years after the generation of trophoblast stem cells in the mouse, this paper is one of three published in 2018 that describe the derivation of human trophoblast stem cells in vitro, paving the way to define the transcription factor networks and mechanistic principles governing human early placental development.
Haider, S. et al. Self-renewing trophoblast organoids recapitulate the developmental program of the early human placenta. Stem Cell Rep. 11, 537–551 (2018). The second of three papers published in 2018 that describe the isolation and perpetuation of human trophoblast stem cells directly from early first-trimester placenta, here using organoid culture approaches.
Turco, M. Y. et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 564, 263–267 (2018). Building on an earlier study by the same group that had generated human endometrial glandular organoids, this is the third of the three papers published in 2018 to describe the derivation of human trophoblast stem cells, in this case through the use of 3D organoid culture systems.
Ramathal, C. Y., Bagchi, I. C., Taylor, R. N. & Bagchi, M. K. Endometrial decidualization: of mice and men. Semin. Reprod. Med. 28, 17–26 (2010).
Knofler, M. Critical growth factors and signalling pathways controlling human trophoblast invasion. Int. J. Dev. Biol. 54, 269–280 (2010).
Moffett, A. & Colucci, F. Uterine NK cells: active regulators at the maternal-fetal interface. J. Clin. Invest. 124, 1872–1879 (2014).
Woods, L. et al. Decidualisation and placentation defects are a major cause of age-related reproductive decline. Nat. Commun. 8, 352 (2017).
Klemmt, P. A., Carver, J. G., Kennedy, S. H., Koninckx, P. R. & Mardon, H. J. Stromal cells from endometriotic lesions and endometrium from women with endometriosis have reduced decidualization capacity. Fertil. Steril. 85, 564–572 (2006).
Copp, A. J. Death before birth: clues from gene knockouts and mutations. Trends Genet. 11, 87–93 (1995).
Perez-Garcia, V. et al. Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 555, 463–468 (2018). This paper describes a comprehensive analysis of placental phenotypes in mouse gene knockouts that cause embryonic lethality, showing that placental development is very frequently affected in these mutants.
Ueno, M. et al. c-Met-dependent multipotent labyrinth trophoblast progenitors establish placental exchange interface. Dev. Cell 27, 373–386 (2013).
Natale, B. V. et al. Sca-1 identifies a trophoblast population with multipotent potential in the mid-gestation mouse placenta. Sci. Rep. 7, 5575 (2017).
James, J. L., Carter, A. M. & Chamley, L. W. Human placentation from nidation to 5 weeks of gestation. Part I: what do we know about formative placental development following implantation? Placenta 33, 327–334 (2012).
Knöfler, M. et al. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell. Mol. Life Sci. https://doi.org/10.1007/s00018-019-03104-6 (2019). This outstanding, comprehensive review of human placental development focuses on stem cells and their use in understanding trophoblast lineage specification and commitment and is accompanied by instructive diagrams and transcription factor hierarchies.
Boyd, J. D. & Hamilton, W. J. The Human Placenta (Heffer & Sons, 1970).
Burton, G. J., Jauniaux, E. & Watson, A. L. Maternal arterial connections to the placental intervillous space during the first trimester of human pregnancy: the Boyd collection revisited. Am. J. Obstet. Gynecol. 181, 718–724 (1999).
Burton, G. J., Watson, A. L., Hempstock, J., Skepper, J. N. & Jauniaux, E. Uterine glands provide histiotrophic nutrition for the human fetus during the first trimester of pregnancy. J. Clin. Endocrinol. Metab. 87, 2954–2959 (2002).
Rossant, J. & Cross, J. C. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2, 538–548 (2001).
Woods, L., Perez-Garcia, V. & Hemberger, M. Regulation of placental development and its impact on fetal growth-new insights from mouse models. Front. Endocrinol. 9, 570 (2018).
Adamson, S. L. et al. Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev. Biol. 250, 358–373 (2002). This seminal study on the architecture of the mouse placental vasculature outlines the anatomy of maternal blood spaces and the nature of maternal–fetal vascular interactions, matching structure to function.
Niwa, H. et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929 (2005).
Arnold, S. J. & Robertson, E. J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009).
Dietrich, J. E. & Hiiragi, T. Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219–4231 (2007).
Strumpf, D. et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 (2005).
Nishioka, N. et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270–283 (2008).
Nishioka, N. et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 (2009).
Blakeley, P. et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3151–3165 (2015).
Horii, M. et al. Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc. Natl Acad. Sci. USA 113, E3882–E3891 (2016).
Soncin, F. et al. Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development 145, dev156273 (2018).
Hemberger, M., Udayashankar, R., Tesar, P., Moore, H. & Burton, G. J. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum. Mol. Genet. 19, 2456–2467 (2010).
Home, P. et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc. Natl Acad. Sci. USA 109, 7362–7367 (2012).
Yagi, R. et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827–3836 (2007).
Kidder, B. L. & Palmer, S. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res. 20, 458–472 (2010).
Latos, P. A. & Hemberger, M. The transcriptional and signalling networks of mouse trophoblast stem cells. Placenta 35, S81–S85 (2014).
Russ, A. P. et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95–99 (2000).
Donnison, M. et al. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development 132, 2299–2308 (2005).
Yamamoto, H. et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 12, 1315–1326 (1998).
Wen, F. et al. Ets2 is required for trophoblast stem cell self-renewal. Dev. Biol. 312, 284–299 (2007).
Luo, J. et al. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature 388, 778–782 (1997).
Adachi, K. et al. Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol. Cell 52, 380–392 (2013).
Latos, P. A. et al. Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nat. Commun. 6, 7776 (2015).
Knott, J. G. & Paul, S. Transcriptional regulators of the trophoblast lineage in mammals with hemochorial placentation. Reproduction 148, R121–R136 (2014).
Ralston, A. et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403 (2010).
Home, P. et al. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J. Biol. Chem. 284, 28729–28737 (2009).
Avilion, A. A. et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 (2003).
Kuckenberg, P. et al. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol. Cell. Biol. 30, 3310–3320 (2010).
Kunath, T. et al. Developmental differences in the expression of FGF receptors between human and mouse embryos. Placenta 35, 1079–1088 (2014).
Lee, C. Q. et al. What is trophoblast? a combination of criteria define human first-trimester trophoblast. Stem Cell Rep. 6, 257–272 (2016).
Lee, Y. et al. A unifying concept of trophoblastic differentiation and malignancy defined by biomarker expression. Hum. Pathol. 38, 1003–1013 (2007).
Niwa, H., Miyazaki, J. & Smith, A. G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 (2000).
Latos, P. A. et al. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev. 29, 2435–2448 (2015).
Hu, S. et al. DNA methylation presents distinct binding sites for human transcription factors. eLife 2, e00726 (2013).
Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017). This paper systematically analyses the DNA-binding preferences of human transcription factors that depend on the methylation state of CpGs contained within their cognate motifs, thus directly linking the transcription factor network with the epigenome to explain cell fate specification and cellular commitment decisions.
Hartl, D. et al. CG dinucleotides enhance promoter activity independent of DNA methylation. Genome Res. 29, 554–563 (2019).
Bourque, G. et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762 (2008).
Pavlicev, M., Hiratsuka, K., Swaggart, K. A., Dunn, C. & Muglia, L. Detecting endogenous retrovirus-driven tissue-specific gene transcription. Genome Biol. Evol. 7, 1082–1097 (2015).
Walsh, C. P., Chaillet, J. R. & Bestor, T. H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117 (1998).
Bestor, T. H. & Bourc'his, D. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb. Symp. Quant. Biol. 69, 381–387 (2004).
Lavie, L., Kitova, M., Maldener, E., Meese, E. & Mayer, J. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79, 876–883 (2005).
Macfarlan, T. S. et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 25, 594–607 (2011).
Cohen, C. J. et al. Placenta-specific expression of the interleukin-2 (IL-2) receptor beta subunit from an endogenous retroviral promoter. J. Biol. Chem. 286, 35543–35552 (2011).
Chuong, E. B., Rumi, M. A., Soares, M. J. & Baker, J. C. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329 (2013). This study identifies endogenous retrovirus-derived LTR elements that serve important functions as trophoblast transcription factor binding sites in mammalian evolution.
Macfarlan, T. S. et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 (2012).
Brind'Amour, J. et al. LTR retrotransposons transcribed in oocytes drive species-specific and heritable changes in DNA methylation. Nat. Commun. 9, 3331 (2018).
Dunn-Fletcher, C. E. et al. Anthropoid primate-specific retroviral element THE1B controls expression of CRH in placenta and alters gestation length. PLOS Biol. 16, e2006337 (2018).
Huh, J. W., Ha, H. S., Kim, D. S. & Kim, H. S. Placenta-restricted expression of LTR-derived NOS3. Placenta 29, 602–608 (2008).
Mi, S. et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 (2000). This paper describes the remarkable situation in which a human endogenous retrovirus-derived gene, HERV-W, has been sequestered to serve an important physiological function in placental syncytiotrophoblast formation.
Dupressoir, A. et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl Acad. Sci. USA 108, E1164–E1173 (2011).
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Dean, W., Santos, F. & Reik, W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin. Cell Dev. Biol. 14, 93–100 (2003).
Seisenberger, S. et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 (2012).
Smallwood, S. A. et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814 (2011).
Hamada, H. et al. Allele-specific methylome and transcriptome analysis reveals widespread imprinting in the human placenta. Am. J. Hum. Genet. 99, 1045–1058 (2016).
Branco, M. R. et al. Maternal DNA methylation regulates early trophoblast development. Dev. Cell 36, 152–163 (2016).
Sanchez-Delgado, M. et al. Human oocyte-derived methylation differences persist in the placenta revealing widespread transient imprinting. PLOS Genet. 12, e1006427 (2016).
Zhang, Y. et al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 50, 96–105 (2018). This is a comprehensive study of transcriptomes and base-resolution DNA methylomes during early-lineage specification in peri- and post-implantation mouse embryos.
Smith, Z. D. et al. Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature 549, 543–547 (2017).
Gamage, T. et al. Human trophoblasts are primarily distinguished from somatic cells by differences in the pattern rather than the degree of global CpG methylation. Biol. Open 7, bio034884 (2018).
Robinson, W. P. & Price, E. M. The human placental methylome. Cold Spring Harb. Perspect. Med. 5, a023044 (2015).
Schroeder, D. I. et al. The human placenta methylome. Proc. Natl Acad. Sci. USA 110, 6037–6042 (2013).
Bianco-Miotto, T. et al. Recent progress towards understanding the role of DNA methylation in human placental development. Reproduction 152, R23–R30 (2016).
Novakovic, B. et al. Wide-ranging DNA methylation differences of primary trophoblast cell populations and derived cell lines: implications and opportunities for understanding trophoblast function. Mol. Hum. Reprod. 17, 344–353 (2011).
Wilson, R. L. et al. Characterization of 5-methylcytosine and 5-hydroxymethylcytosine in human placenta cell types across gestation. Epigenetics 14, 660–671 (2019).
Luo, C., Hajkova, P. & Ecker, J. R. Dynamic DNA methylation: in the right place at the right time. Science 361, 1336–1340 (2018).
Decato, B. E., Lopez-Tello, J., Sferruzzi-Perri, A. N., Smith, A. D. & Dean, M. D. DNA methylation divergence and tissue specialization in the developing mouse placenta. Mol. Biol. Evol. 34, 1702–1712 (2017).
Mohammed, H. et al. Single-cell landscape of transcriptional heterogeneity and cell fate decisions during mouse early gastrulation. Cell Rep. 20, 1215–1228 (2017).
Rulands, S. et al. Genome-scale oscillations in DNA methylation during exit from pluripotency. Cell Syst. 7, 63–76.e12 (2018).
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011).
Senner, C. E., Krueger, F., Oxley, D., Andrews, S. & Hemberger, M. DNA methylation profiles define stem cell identity and reveal a tight embryonic-extraembryonic lineage boundary. Stem Cells 30, 2732–2745 (2012).
Khoueiry, R. et al. Lineage-specific functions of TET1 in the postimplantation mouse embryo. Nat. Genet. 49, 1061–1072 (2017).
Chrysanthou, S. et al. A critical role of TET1/2 proteins in cell-cycle progression of trophoblast stem cells. Stem Cell Rep. 10, 1355–1368 (2018).
Dawlaty, M. M. et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175 (2011).
Schoenfelder, S. et al. Divergent wiring of repressive and active chromatin interactions between mouse embryonic and trophoblast lineages. Nat. Commun. 9, 4189 (2018).
Fogarty, N. M., Burton, G. J. & Ferguson-Smith, A. C. Different epigenetic states define syncytiotrophoblast and cytotrophoblast nuclei in the trophoblast of the human placenta. Placenta 36, 796–802 (2015).
Ng, R. K. et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 10, 1280–1290 (2008). This study identifies the transcription factor Elf5 as an important broker of the first cell fate decision by epigenetic repression in the embryonic lineage, which provides a pioneering molecular example underpinning Waddington’s model of the canalization of developmental pathways.
Dodge, J. E., Kang, Y. K., Beppu, H., Lei, H. & Li, E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 24, 2478–2486 (2004).
Yeap, L. S., Hayashi, K. & Surani, M. A. ERG-associated protein with SET domain (ESET)–Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin 2, 12 (2009).
Yuan, P. et al. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 23, 2507–2520 (2009).
Tachibana, M. et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 (2002).
O'Carroll, D. et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21, 4330–4336 (2001).
Pasini, D., Bracken, A. P., Jensen, M. R., Lazzerini Denchi, E. & Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23, 4061–4071 (2004).
Rugg-Gunn, P. J., Cox, B. J., Ralston, A. & Rossant, J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc. Natl Acad. Sci. USA 107, 10783–10790 (2010).
Alder, O. et al. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development 137, 2483–2492 (2010).
Lewis, A. et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 36, 1291–1295 (2004). This landmark study highlights a lesser, albeit not insignificant, reliance of trophoblast cells on DNA methylation than in embryonic tissues.
Umlauf, D. et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 36, 1296–1300 (2004).
Wagschal, A. & Feil, R. Genomic imprinting in the placenta. Cytogenet. Genome Res. 113, 90–98 (2006).
Sakaue, M. et al. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr. Biol. 20, 1452–1457 (2010).
Yang, X. et al. Silencing of developmental genes by H3K27me3 and DNA methylation reflects the discrepant plasticity of embryonic and extraembryonic lineages. Cell Res. 28, 593–596 (2018).
Nugent, B. M., O'Donnell, C. M., Epperson, C. N. & Bale, T. L. Placental H3K27me3 establishes female resilience to prenatal insults. Nat. Commun. 9, 2555 (2018).
Inoue, A., Jiang, L., Lu, F., Suzuki, T. & Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 (2017).
Hanna, C. W. et al. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 26, 756–767 (2016).
Xu, J. & Kidder, B. L. KDM5B decommissions the H3K4 methylation landscape of self-renewal genes during trophoblast stem cell differentiation. Biol. Open 7, bio031245 (2018).
Fedorov, O. et al. Selective targeting of the BRG/PB1 bromodomains impairs embryonic and trophoblast stem cell maintenance. Sci. Adv. 1, e1500723 (2015).
Paauw, N. D. et al. H3K27 acetylation and gene expression analysis reveals differences in placental chromatin activity in fetal growth restriction. Clin. Epigenetics 10, 85 (2018).
Hepp, P. et al. Histone H3 lysine 9 acetylation is downregulated in GDM placentas and calcitriol supplementation enhanced this effect. Int. J. Mol. Sci. 19, 4061 (2018).
Lee, E. Y. et al. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359, 288–294 (1992).
Clarke, A. R. et al. Requirement for a functional Rb-1 gene in murine development. Nature 359, 328–330 (1992).
Davis, A. C., Wims, M., Spotts, G. D., Hann, S. R. & Bradley, A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7, 671–682 (1993).
Wu, L. et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421, 942–947 (2003).
Dubois, N. C. et al. Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development 135, 2455–2465 (2008).
Ayadi, A. et al. Mouse large-scale phenotyping initiatives: overview of the European Mouse Disease Clinic (EUMODIC) and of the wellcome trust sanger institute mouse genetics project. Mamm. Genome 23, 600–610 (2012).
de Angelis, M. H. et al. Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat. Genet. 47, 969–978 (2015).
White, J. K. et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154, 452–464 (2013).
Dickinson, M. E. et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016).
Adams, R. H. et al. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol. Cell 6, 109–116 (2000).
Barak, Y. et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell 4, 585–595 (1999). This study is one of the first to highlight the connection between placenta and heart development and to prove that a defective placenta can be the sole cause of cardiac defects in the mouse.
O'Keeffe, G. W. & Kenny, L. C. Predicting infant neurodevelopmental outcomes using the placenta? Trends Mol. Med. 20, 303–305 (2014).
Hodyl, N. A. et al. Child neurodevelopmental outcomes following preterm and term birth: what can the placenta tell us? Placenta 57, 79–86 (2017).
Maltepe, E. & Fisher, S. J. Placenta: the forgotten organ. Annu. Rev. Cell Dev. Biol. 31, 523–552 (2015).
Bonnin, A. et al. A transient placental source of serotonin for the fetal forebrain. Nature 472, 347–350 (2011). This study identifies that, in both mouse and humans, the placenta acts as a neuroendocrine organ by producing serotonin, a molecule critical for neurodevelopment in the fetus, thus offering a mechanism connecting placental insufficiency with diminished postnatal cognitive abilities and later-life mental health issues.
Ho, L. et al. ELABELA deficiency promotes preeclampsia and cardiovascular malformations in mice. Science 357, 707–713 (2017).
Beccari, L. et al. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562, 272–276 (2018).
Jones, H. N. et al. Hypoplastic left heart syndrome is associated with structural and vascular placental abnormalities and leptin dysregulation. Placenta 36, 1078–1086 (2015).
Matthiesen, N. B. et al. Congenital heart defects and indices of placental and fetal growth in a nationwide study of 924 422 liveborn infants. Circulation 134, 1546–1556 (2016).
Rychik, J. et al. Characterization of the placenta in the newborn with congenital heart disease: distinctions based on type of cardiac malformation. Pediatr. Cardiol. 39, 1165–1171 (2018).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Yoney, A. et al. WNT signaling memory is required for ACTIVIN to function as a morphogen in human gastruloids. eLife 7, e38279 (2018).
Fogarty, N. M. E. et al. Genome editing reveals a role for OCT4 in human embryogenesis. Nature 550, 67–73 (2017).
Sozen, B. et al. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 20, 979–989 (2018). This study highlights that mouse gastrulation stage embryo-like structures can self-assemble from the three component stem cell populations (that is, embryonic, extra-embryonic endoderm and trophoblast stem cells) under specific conditions, thereby creating the possibility of studying very early post-implantation-like stages in vitro.
Harrison, S. E., Sozen, B., Christodoulou, N., Kyprianou, C. & Zernicka-Goetz, M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356, eaal1810 (2017).
Rivron, N. C. et al. Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111 (2018). This detailed study demonstrates that the assembly of embryonic and trophoblast stem cells in optimal ratios enables the formation of blastocyst-like structures that provide an ideal tool to study embryonic–trophoblast cell interactions.
Rivron, N. et al. Debate ethics of embryo models from stem cells. Nature 564, 183–185 (2018).
Turco, M. Y. et al. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 19, 568–577 (2017).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Hata, K., Okano, M., Lei, H. & Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 (2002).
Reik, W., Dean, W. & Walter, J. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 (2001).
Nikitina, T. et al. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol. Cell. Biol. 27, 864–877 (2007).
Maurano, M. T. et al. Role of DNA methylation in modulating transcription factor occupancy. Cell Rep. 12, 1184–1195 (2015).
Dean, W. DNA methylation and demethylation: a pathway to gametogenesis and development. Mol. Reprod. Dev. 81, 113–125 (2014).
Lister, R. et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013).
Gabel, H. W. et al. Disruption of DNA-methylation-dependent long gene repression in rett syndrome. Nature 522, 89–93 (2015).
Lucas, E. S. et al. Loss of endometrial plasticity in recurrent pregnancy loss. Stem Cells 34, 346–356 (2016).
Knofler, M. et al. Human Hand1 basic helix-loop-helix (bHLH) protein: extra-embryonic expression pattern, interaction partners and identification of its transcriptional repressor domains. Biochem. J. 361, 641–651 (2002).
Donnison, M., Broadhurst, R. & Pfeffer, P. L. Elf5 and Ets2 maintain the mouse extraembryonic ectoderm in a dosage dependent synergistic manner. Dev. Biol. 397, 77–88 (2015).
Hu, Y. G. et al. Regulation of DNA methylation activity through Dnmt3L promoter methylation by Dnmt3 enzymes in embryonic development. Hum. Mol. Genet. 17, 2654–2664 (2008).
Etoc, F. et al. A balance between secreted inhibitors and edge sensing controls gastruloid self-organization. Dev. Cell 39, 302–315 (2016).
Fonseca, B. M., Correia-da-Silva, G. & Teixeira, N. A. The rat as an animal model for fetoplacental development: a reappraisal of the post-implantation period. Reprod. Biol. 12, 97–118 (2012).
Soares, M. J., Chakraborty, D., Karim Rumi, M. A., Konno, T. & Renaud, S. J. Rat placentation: an experimental model for investigating the hemochorial maternal–fetal interface. Placenta 33, 233–243 (2012).
Sahgal, N., Canham, L. N., Canham, B. & Soares, M. J. Rcho-1 trophoblast stem cells: a model system for studying trophoblast cell differentiation. Methods Mol. Med. 121, 159–178 (2006).
Swanson, A. M. & David, A. L. Animal models of fetal growth restriction: considerations for translational medicine. Placenta 36, 623–630 (2015).
Andersen, M. D. et al. in Experimental Animal Models of Human Diseases (ed. B. Ibeh). (IntechOpen, 2018).
Grigsby, P. L. Animal models to study placental development and function throughout normal and dysfunctional human pregnancy. Semin. Reprod. Med. 34, 11–16 (2016).
Fischer, B., Chavatte-Palmer, P., Viebahn, C., Navarrete Santos, A. & Duranthon, V. Rabbit as a reproductive model for human health. Reproduction 144, 1–10 (2012).
Tan, T. et al. Generation of trophoblast stem cells from rabbit embryonic stem cells with BMP4. PLOS ONE 6, e17124 (2011).
Handwerger, S. et al. Development of the sheep as an animal model to study placental lactogen physiology. J. Pediatr. 87, 1139–1143 (1975).
Barry, J. S. & Anthony, R. V. The pregnant sheep as a model for human pregnancy. Theriogenology 69, 55–67 (2008).
Orendi, K. et al. Placental and trophoblastic in vitro models to study preventive and therapeutic agents for preeclampsia. Placenta 32, S49–S54 (2011).
Newby, D., Marks, L., Cousins, F., Duffie, E. & Lyall, F. Villous explant culture: characterization and evaluation of a model to study trophoblast invasion. Hypertens. Pregnancy 24, 75–91 (2005).
Sooranna, S. R., Oteng-Ntim, E., Meah, R., Ryder, T. A. & Bajoria, R. Characterization of human placental explants: morphological, biochemical and physiological studies using first and third trimester placenta. Hum. Reprod. 14, 536–541 (1999).
Pattillo, R. A. & Gey, G. O. The establishment of a cell line of human hormone-synthesizing trophoblastic cells in vitro. Cancer Res. 28, 1231–1236 (1968).
Pattillo, R. A., Gey, G. O., Delfs, E. & Mattingly, R. F. Human hormone production in vitro. Science 159, 1467–1469 (1968).
Kohler, P. O. & Bridson, W. E. Isolation of hormone-producing clonal lines of human choriocarcinoma. J. Clin. Endocrinol. Metab. 32, 683–687 (1971).
Hannan, N. J., Paiva, P., Dimitriadis, E. & Salamonsen, L. A. Models for study of human embryo implantation: choice of cell lines? Biol. Reprod. 82, 235–245 (2010).
Graham, C. H. et al. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp. Cell Res. 206, 204–211 (1993).
Choy, M. Y. & Manyonda, I. T. The phagocytic activity of human first trimester extravillous trophoblast. Hum. Reprod. 13, 2941–2949 (1998).
King, A., Thomas, L. & Bischof, P. Cell culture models of trophoblast II: trophoblast cell lines — a workshop report. Placenta 21, S113–S119 (2000).
Whitley, G. S. Production of human trophoblast cell lines. Methods Mol. Med. 121, 219–228 (2006).
Xu, R. H. et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264 (2002).
Zdravkovic, T. et al. Human stem cells from single blastomeres reveal pathways of embryonic or trophoblast fate specification. Development 142, 4010–4025 (2015).
Acknowledgements
The authors’ programme of work is funded by a Next Generation Fellowship from the Centre for Trophoblast Research to C.W.H.; by a Tier I Canada Research Chair grant to M.H.; by the Magee Prize, funded by the Richard King Mellon Foundation; and by the Alberta Children’s Hospital Research Institute.
Reviewer information
Nature Reviews Genetics thanks S. Paul, J. Rossant and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
W.D. and M.H. contributed to discussion of the content. All authors researched data for the article. The bulk of the article was written by M.H. and W.D., with essential contributions on human development and global epigenome studies by C.W.H. All authors reviewed and/or edited the article before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Trophoblast
-
The early cell lineage that arises at the blastocyst stage and gives rise to most extra-embryonic tissues, most notably those of the placenta.
- Myometrium
-
The middle layer of the uterine wall, which consists of smooth muscle cells that are important for contraction.
- Allogeneic
-
The phenomenon of cells or tissues being genetically different and hence distinct from each other in terms of cell surface antigen repertoire, leading to immunological incompatibility.
- Haemochorial placentation
-
The anatomical arrangement in placentas in which trophoblast-lined villi are directly exposed to maternal blood.
- Decidualization
-
The process by which endometrial stromal fibroblasts become specialized secretory decidual cells during pregnancy.
- Chromatin
-
The complex of DNA, RNA and proteins (mostly histones) that collectively comprise the chromosomes.
- Epigenome
-
The sum total of all modifications to DNA, or to DNA-associated RNA and proteins, that permit interpretation of the genome to instruct cell identity and function.
- Syncytialization
-
The process of cell–cell fusion that leads to the formation of a multi-nucleate cell. It occurs when cytotrophoblast cells fuse during placental development to form syncytiotrophoblast.
- Imprinted genes
-
The relatively small collection of genes whose expression is solely based on parent-of-origin inheritance of the allele.
- Epimutations
-
Changes in the modification status of a cytosine (that is, DNA methylation, hydroxymethylation or derivatives), usually found in the context of CpG dinucleotides, between successive cell divisions or generations.
- Transgenerational inheritance
-
Inheritance across multiple successive (>2) generations of instructive elements of the epigenome, found in germ cells (oocytes or sperm).
- CpG islands
-
(CGIs). Regions of the genome in which the CpG frequency is greater than 50% GC content over at least 200 base pairs.
Rights and permissions
About this article
Cite this article
Hemberger, M., Hanna, C.W. & Dean, W. Mechanisms of early placental development in mouse and humans. Nat Rev Genet 21, 27–43 (2020). https://doi.org/10.1038/s41576-019-0169-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-019-0169-4
This article is cited by
-
Paternal DDT exposure induces sex-specific programming of fetal growth, placenta development and offspring’s health phenotypes in a mouse model
Scientific Reports (2024)
-
Transmission, seroprevalence, and maternal-fetal impact of lymphocytic choriomeningitis virus
Pediatric Research (2024)
-
Single-nucleus multi-omic profiling of human placental syncytiotrophoblasts identifies cellular trajectories during pregnancy
Nature Genetics (2024)
-
The ‘communicatome’ of pregnancy: spotlight on cellular and extravesicular chimerism
EMBO Molecular Medicine (2024)
-
Effect of SARS-CoV-2 infection in early pregnancy on placental development
Science China Life Sciences (2024)