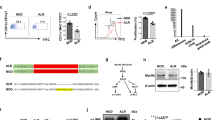
Abstract
Genome-wide association studies have implicated more than 50 genomic regions in type 1 diabetes (T1D). A T1D region at chromosome 16p13.13 includes the candidate genes CLEC16A and DEXI. Conclusive evidence as to which gene is causal for the disease association of this region is missing. We previously reported that Clec16a deficiency modified immune reactivity and protected against autoimmunity in the nonobese diabetic (NOD) mouse model for T1D. However, the diabetes-associated SNPs at 16p13.13 were described to also impact on DEXI expression and others have argued that DEXI is the causal gene in this disease locus. To help resolve whether DEXI affects disease, we generated Dexi knockout (KO) NOD mice. We found that Dexi deficiency had no effect on the frequency of diabetes. To test for possible interactions between Dexi and Clec16a, we intercrossed Dexi KO and Clec16a knockdown (KD) NOD mice. Dexi KO did not modify the disease protection afforded by Clec16a KD. We conclude that Dexi plays no role in autoimmune diabetes in the NOD model. Our data provide strongly suggestive evidence that CLEC16A, not DEXI, is causal for the T1D association of variants in the 16p13.13 region.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–6.
Tomlinson MJ 4th, Pitsillides A, Pickin R, Mika M, Keene KL, Hou X, et al. Fine mapping and functional studies of risk variants for type 1 diabetes at chromosome 16p13.13. Diabetes. 2014;63:4360–8.
WellCome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78.
Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–4.
Mero IL, Ban M, Lorentzen ÅR, Smestad C, Celius EG, Sæther H, et al. Exploring the CLEC16A gene reveals a MS-associated variant with correlation to the relative expression of CLEC16A isoforms in thymus. Genes Immun. 2011;12:191–8.
Leikfoss IS, Keshari PK, Gustavsen MW, Bjolgerud A, Brorson IS, Celius EG, et al. Multiple sclerosis risk allele in CLEC16A acts as an expression quantitative trait locus for CLEC16A and SOCS1 in CD4+ T cells. PLoS ONE. 2015;10:e0132957.
Schuster C, Gerold KD, Schober K, Probst L, Boerner K, Kim MJ, et al. The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity. 2015;42:942–52.
Davison LJ, Wallace C, Cooper JD, Cope NF, Wilson NK, Smyth DJ, et al. Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene. Hum Mol Genet. 2012;21:322–33.
Leikfoss IS, Mero IL, Dahle MK, Lie BA, Harbo HF, Spurkland A, et al. Multiple sclerosis-associated single-nucleotide polymorphisms in CLEC16A correlate with reduced SOCS1 and DEXI expression in the thymus. Genes Immun. 2013;14:62–66.
Dos Santos RS, Marroqui L, Velayos T, Olazagoitia-Garmendia A, Jauregi-Miguel A, Castellanos-Rubio A, et al. DEXI, a candidate gene for type 1 diabetes, modulates rat and human pancreatic beta cell inflammation via regulation of the type I IFN/STAT signalling pathway. Diabetologia. 2019;62:459–72.
Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–57.
Yang H, Wang H, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc. 2014;9:1956–68.
Langevin C, Aleksejeva E, Passoni G, Palha N, Levraud JP, Boudinot P. The antiviral innate immune response in fish: evolution and conservation of the IFN system. J Mol Biol. 2013;425:4904–20.
Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J, et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell. 2014;157:1577–90.
Davison LJ, Wallace MD, Preece C, Hughes K, Todd JA, Davies B, et al. Dexi disruption depletes gut microbial metabolites and accelerates autoimmune diabetes. bioRxiv 393421; https://doi.org/10.1101/393421.
Acknowledgements
We thank John Stockton for embryo microinjections, and also thank Charles Evavold for advice on BM-DM experiments and providing conditioned media. JMNB was supported by a pre-doctoral training fellowship from NIDDK (T32DK007260). This research was funded in part by a grant from NIDDK (R56DK109954-01) to SK and supported by core facilities funded by the NIDDK Diabetes Research Center award P30DK036836 to the Joslin Diabetes Center.
Author information
Authors and Affiliations
Contributions
JMNB performed experiments, analyzed data, and wrote the manuscript. BK performed experiments. CS helped with experimental design and data interpretation. SK supervised the study, analyzed data, and wrote the manuscript. SK is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nieves-Bonilla, J.M., Kiaf, B., Schuster, C. et al. The type 1 diabetes candidate gene Dexi does not affect disease risk in the nonobese diabetic mouse model. Genes Immun 21, 71–77 (2020). https://doi.org/10.1038/s41435-019-0083-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-019-0083-y