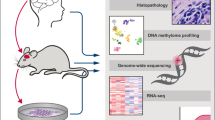
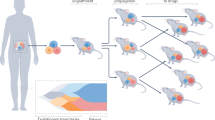
Abstract
The spectrum of tumours arising in childhood is fundamentally different from that seen in adults, and they are known to be divergent from adult malignancies in terms of cellular origins, epidemiology, genetic complexity, driver mutations and underlying mutational processes. Despite the immense knowledge generated through sequencing efforts and functional characterization of identified (epi-)genetic alterations over the past decade, the clinical implications of this knowledge have so far been limited. Novel preclinical platforms such as the European Innovative Therapies for Children with Cancer–Paediatric Preclinical Proof-of-Concept Platform and the US-based Pediatric Preclinical Testing Consortium are being developed to try to change this by aiming to recapitulate the extensive heterogeneity of paediatric tumours and thereby, hopefully, improve the ability to predict clinical benefit. Numerous studies have also been established worldwide to provide patients with access to real-time molecular profiling and the possibility of more precise mechanism-of-action-based treatments. In addition to tumour-intrinsic findings and mechanisms, ongoing studies are investigating features such as the immune microenvironment of paediatric tumours in comparison with adult cancers — currently of very timely clinical relevance. However, there is an ongoing need for rigorous preclinical biomarker and target validation to feed into the next generation of molecularly stratified clinical trials. This Review aims to provide a comprehensive state-of-the-art overview of the molecular landscape of paediatric solid tumours, including their underlying genomic alterations and interactions with the microenvironment, complemented with our current understanding of potential therapeutic vulnerabilities and how these can be preclinically tested using more accurate predictive methods. Finally, we provide an outlook on the challenges and opportunities associated with translating this overwhelming scientific progress into real clinical benefit.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Grobner, S. N. et al. The landscape of genomic alterations across childhood cancers. Nature 555, 321–327 (2018).
Ma, X. et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–376 (2018). Together with Grobner et al., this study describes the largest pan-cancer genomic analyses of paediatric cancer to date, with information on patterns of genetic alterations, drug targets and other molecular features.
Brodeur, G. M., Nichols, K. E., Plon, S. E., Schiffman, J. D. & Malkin, D. Pediatric cancer predisposition and surveillance: an overview, and a tribute to Alfred G. Knudson Jr. Clin. Cancer Res. 23, e1–e5 (2017).
Zhang, J. et al. Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med. 373, 2336–2346 (2015).
Huang, K. L. et al. Pathogenic germline variants in 10,389 adult cancers. Cell 173, 355–370 (2018).
Linabery, A. M. & Ross, J. A. Trends in childhood cancer incidence in the U.S. (1992–2004). Cancer 112, 416–432 (2008).
Ostrom, Q. T. et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro-oncology 20, iv1–iv86 (2018).
Jones, D. T. et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat. Genet. 45, 927–932 (2013).
Zhang, J. et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 45, 602–612 (2013).
Ramkissoon, L. A. et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc. Natl Acad. Sci. USA 110, 8188–8193 (2013).
Bandopadhayay, P. et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat. Genet. 48, 273–282 (2016).
Jones, C. & Baker, S. J. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat. Rev. Cancer 14, 651–661 (2014).
Mackay, A. et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 32, 520–537 (2017).
Sturm, D. et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat. Rev. Cancer 14, 92–107 (2014).
Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 (2009).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Wu, G. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 (2012). Together with Schwartzentruber et al., this study describes, for the first time, the widespread occurrence of mutations in histone genes in paediatric high-grade gliomas.
Buczkowicz, P. et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat. Genet. 46, 451–456 (2014).
Fontebasso, A. M. et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat. Genet. 46, 462–466 (2014).
Taylor, K. R. et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat. Genet. 46, 457–461 (2014).
Wu, G. et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat. Genet. 46, 444–450 (2014).
Lange, A. M. & Lo, H. W. Inhibiting TRK proteins in clinical cancer therapy. Cancers (Basel) 10, (E105 (2018).
Pajtler, K. W. et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27, 728–743 (2015).
Pajtler, K. W. et al. Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 136, 211–226 (2018).
Panwalkar, P. et al. Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol. 134, 705–714 (2017).
Bayliss, J. et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci. Transl Med. 8, 366ra161 (2016).
Hubner, J. M. et al. EZHIP/CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro-oncology https://doi.org/10.1093/neuonc/noz058 (2019).
Jain, S. U. et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat. Commun. 10, 2146 (2019).
World Health Organization. WHO Classification of Tumours of the Central Nervous System, Revised. Vol. 1 4th edn (eds Louis, D. N., Ohgaki, H., Wiestler, O. D. & Cavenee, W. K.) (IARC, 2016).
Taylor, M. D. et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 123, 465–472 (2012).
Northcott, P. A. et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017).
Northcott, P. A. et al. Medulloblastoma. Nat. Rev. Dis. Primers 5, 11 (2019).
Cavalli, F. M. G. et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31, 737–754 (2017).
Schwalbe, E. C. et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 18, 958–971 (2017).
Robinson, G. W. et al. Risk-adapted therapy for young children with medulloblastoma (SJYC07): therapeutic and molecular outcomes from a multicentre, phase 2 trial. Lancet Oncol. 19, 768–784 (2018).
Versteege, I. et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998).
Johann, P. D. et al. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell 29, 379–393 (2016).
Torchia, J. et al. Integrated (epi)-genomic analyses identify subgroup-specific therapeutic targets in CNS rhabdoid tumors. Cancer Cell 30, 891–908 (2016).
Hasselblatt, M. et al. Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. Am. J. Surg. Pathol. 35, 933–935 (2011).
Schneppenheim, R. et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am. J. Hum. Genet. 86, 279–284 (2010).
Sturm, D. et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164, 1060–1072 (2016). This paper uses molecular classification methods to show that the majority of what was previously termed CNS-PNETs are, in fact, other known tumour entities, and also identified four novel tumour classes with distinct features.
Hwang, E. I. et al. Extensive molecular and clinical heterogeneity in patients with histologically diagnosed CNS-PNET treated as a single entity: a report from the Children’s Oncology Group randomized ACNS0332 trial. J. Clin. Oncol. 34, 3388–3395 (2018).
Hingorani, P. et al. Current state of pediatric sarcoma biology and opportunities for future discovery: a report from the sarcoma translational research workshop. Cancer Genet. 209, 182–194 (2016).
Martin, J. W., Squire, J. A. & Zielenska, M. The genetics of osteosarcoma. Sarcoma 2012, 627254 (2012).
Grunewald, T. G. P. et al. Ewing sarcoma. Nat. Rev. Dis. Primers 4, 5 (2018).
Sorensen, P. H. et al. A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat. Genet. 6, 146–151 (1994).
Delattre, O. et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359, 162–165 (1992). This paper describes, for the first time, the EWSR1 – FLI1 gene fusion, which is pathognomonic of Ewing sarcoma.
Crompton, B. D. et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 4, 1326–1341 (2014).
Tirode, F. et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 4, 1342–1353 (2014).
Kawamura-Saito, M. et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 15, 2125–2137 (2006).
Watson, S. et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J. Pathol. 245, 29–40 (2018).
Pierron, G. et al. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat. Genet. 44, 461–466 (2012). This paper describes, for the first time, BCOR – CCNB3 translocated sarcomas that were identified through a systematic RNA-seq approach on hitherto unclassified EWSR1–ETS-negative small round cell sarcomas.
Szuhai, K. et al. The NFATc2 gene is involved in a novel cloned translocation in a Ewing sarcoma variant that couples its function in immunology to oncology. Clin. Cancer Res. 15, 2259–2268 (2009).
Renzi, S., Anderson, N. D., Light, N. & Gupta, A. Ewing-like sarcoma: an emerging family of round cell sarcomas. J. Cell. Physiol. 234, 7999–8007 (2019).
Baldauf, M. C. et al. Are EWSR1-NFATc2-positive sarcomas really Ewing sarcomas? Mod. Pathol. 31, 997–999 (2018).
Galili, N. et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 5, 230–235 (1993).
Shapiro, D. N., Sublett, J. E., Li, B., Downing, J. R. & Naeve, C. W. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 53, 5108–5112 (1993). Together with Galili et al., this study is the first to describe the PAX – FOXO fusion, which is pathognomonic of alveolar rhabdomyosarcoma.
Koufos, A. et al. Loss of heterozygosity in three embryonal tumours suggests a common pathogenetic mechanism. Nature 316, 330–334 (1985).
Agaram, N. P. et al. Recurrent MYOD1 mutations in pediatric and adult sclerosing and spindle cell rhabdomyosarcomas: evidence for a common pathogenesis. Genes Chromosomes Cancer 53, 779–787 (2014).
Shern, J. F. et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 4, 216–231 (2014).
Spunt, S. L., Skapek, S. X. & Coffin, C. M. Pediatric nonrhabdomyosarcoma soft tissue sarcomas. Oncologist 13, 668–678 (2008).
Waxweiler, T. V. et al. Non-rhabdomyosarcoma soft tissue sarcomas in children: a surveillance, epidemiology, and end results analysis validating COG risk stratifications. Int. J. Radiat. Oncol. Biol. Phys. 92, 339–348 (2015).
Mertens, F., Antonescu, C. R. & Mitelman, F. Gene fusions in soft tissue tumors: recurrent and overlapping pathogenetic themes. Genes Chromosomes Cancer 55, 291–310 (2016).
Clark, J. et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat. Genet. 7, 502–508 (1994). This paper describes, for the first time, the SS18 – SSX gene fusion, which is pathognomonic of synovial sarcoma.
Ladanyi, M. et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene 20, 48–57 (2001).
Crozat, A., Aman, P., Mandahl, N. & Ron, D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363, 640–644 (1993).
Knezevich, S. R., McFadden, D. E., Tao, W., Lim, J. F. & Sorensen, P. H. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat. Genet. 18, 184–187 (1998). This paper describes, for the first time, the ETV6 – NTRK3 fusion in congenital fibrosarcoma, which laid the foundation for the development of NTRK inhibitors and their use in NTRK-fusion-expressing tumours.
Ladanyi, M. & Gerald, W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 54, 2837–2840 (1994).
Matthay, K. K. et al. Neuroblastoma. Nat. Rev. Dis. Primers 2, 16078 (2016).
Schwab, M., Westermann, F., Hero, B. & Berthold, F. Neuroblastoma: biology and molecular and chromosomal pathology. Lancet Oncol. 4, 472–480 (2003).
Janoueix-Lerosey, I. et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455, 967–970 (2008).
Mosse, Y. P. et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455, 930–935 (2008).
Cheung, N. K. et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 307, 1062–1071 (2012).
Molenaar, J. J. et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483, 589–593 (2012).
Peifer, M. et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526, 700–704 (2015).
Ackermann, S. et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science 362, 1165–1170 (2018).
Dunn, J. M., Phillips, R. A., Becker, A. J. & Gallie, B. L. Identification of germline and somatic mutations affecting the retinoblastoma gene. Science 241, 1797–1800 (1988). This paper presents the first identification of somatic and germline mutations in the RB1 gene as a defining feature of retinoblastoma.
Rushlow, D. E. et al. Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet Oncol. 14, 327–334 (2013).
Zhang, J. et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481, 329–334 (2012).
Kooi, I. E. et al. Somatic genomic alterations in retinoblastoma beyond RB1 are rare and limited to copy number changes. Sci. Rep. 6, 25264 (2016).
Laurie, N. A. et al. Inactivation of the p53 pathway in retinoblastoma. Nature 444, 61–66 (2006).
Zielinski, B. et al. Detection of chromosomal imbalances in retinoblastoma by matrix-based comparative genomic hybridization. Genes Chromosomes Cancer 43, 294–301 (2005).
Dimaras, H. et al. Retinoblastoma. Nat. Rev. Dis. Primers 1, 15021 (2015).
Lee, J. S., Sanchez, T. R. & Wootton-Gorges, S. Malignant renal tumors in children. J. Kidney Cancer VHL 2, 84–89 (2015).
Torrezan, G. T. et al. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat. Commun. 5, 4039 (2014).
Walz, A. L. et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 27, 286–297 (2015).
Wegert, J. et al. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell 27, 298–311 (2015).
Rivera, M. N. et al. An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science 315, 642–645 (2007).
Gadd, S. et al. A Children’s Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat. Genet. 49, 1487–1494 (2017).
Astolfi, A. et al. Whole transcriptome sequencing identifies BCOR internal tandem duplication as a common feature of clear cell sarcoma of the kidney. Oncotarget 6, 40934–40939 (2015).
Roy, A. et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat. Commun. 6, 8891 (2015).
Ueno-Yokohata, H. et al. Consistent in-frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat. Genet. 47, 861–863 (2015).
Scott, R. H. et al. Surveillance for Wilms tumour in at-risk children: pragmatic recommendations for best practice. Arch. Dis. Child 91, 995–999 (2006).
French, C. NUT midline carcinoma. Nat. Rev. Cancer 14, 149–150 (2014).
Ripperger, T. et al. Childhood cancer predisposition syndromes — a concise review and recommendations by the Cancer Predisposition Working Group of the Society for Pediatric Oncology and Hematology. Am. J. Med. Genet. A 173A, 1017–1037 (2017).
McBride, K. A. et al. Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat. Rev. Clin. Oncol. 11, 260–271 (2014).
Agaimy, A. & Foulkes, W. D. Hereditary SWI/SNF complex deficiency syndromes. Semin. Diagn. Pathol. 35, 193–198 (2018).
Kuhlen, M. et al. Family-based germline sequencing in children with cancer. Oncogene 38, 1367–1380 (2019).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Schulte, J. H. et al. High ALK receptor tyrosine kinase expression supersedes ALK mutation as a determining factor of an unfavorable phenotype in primary neuroblastoma. Clin. Cancer Res. 17, 5082–5092 (2011).
Mosse, Y. P. et al. Targeting ALK with crizotinib in pediatric anaplastic large cell lymphoma and inflammatory myofibroblastic tumor: a Children’s Oncology Group Study. J. Clin. Oncol. 35, 3215–3221 (2017).
Li, S. Q. et al. Targeting wild-type and mutationally activated FGFR4 in rhabdomyosarcoma with the inhibitor ponatinib (AP24534). PLOS ONE 8, e76551 (2013).
Albert, C. M., Davis, J. L., Federman, N., Casanova, M. & Laetsch, T. W. TRK fusion cancers in children: a clinical review and recommendations for screening. J. Clin. Oncol. 37, 513–524 (2019).
Okamura, R. et al. Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis. Oncol. https://doi.org/10.1200/PO.18.00183 (2018).
Laetsch, T. W. et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol. 19, 705–714 (2018). This paediatric study shows an unprecedented objective response rate of 93% for patients with TRK fusion-positive cancers to the NTRK inhibitor larotrectinib, without major toxicities.
Broniscer, A. et al. Phase 1 trial, pharmacokinetics, and pharmacodynamics of dasatinib combined with crizotinib in children with recurrent or progressive high-grade and diffuse intrinsic pontine glioma. Pediatr. Blood Cancer 65, e27035 (2018).
Bender, S. et al. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat. Med. 22, 1314–1320 (2016).
Korshunov, A. et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 134, 507–516 (2017).
Eleveld, T. F. et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat. Genet. 47, 864–871 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02684058 (2019).
Banerjee, A. et al. A phase I trial of the MEK inhibitor selumetinib (AZD6244) in pediatric patients with recurrent or refractory low-grade glioma: a Pediatric Brain Tumor Consortium (PBTC) study. Neuro-oncology 19, 1135–1144 (2017).
Dombi, E. et al. Activity of selumetinib in neurofibromatosis type 1-related plexiform neurofibromas. N. Engl. J. Med. 375, 2550–2560 (2016). This study shows that paediatric patients with NF1 and inoperable plexiform neurofibromas benefited from treatment with the MEK inhibitor selumetinib, with partial responses in 71% of patients in the absence of excess toxic effects.
van Tilburg, C. M. et al. Response in a child with a BRAF V600E mutated desmoplastic infantile astrocytoma upon retreatment with vemurafenib. Pediatr. Blood Cancer 65, e26893 (2018).
Curigliano, G. & Shah, R. R. Safety and tolerability of phosphatidylinositol-3-kinase (PI3K) inhibitors in oncology. Drug Saf. 42, 247–262 (2019).
Rodon, J., Dienstmann, R., Serra, V. & Tabernero, J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat. Rev. Clin. Oncol. 10, 143–153 (2013).
Krueger, D. A. et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N. Engl. J. Med. 363, 1801–1811 (2010). This study shows that treatment with the mTOR inhibitor everolimus leads to a marked reduction in the volume of subependymal giant-cell astrocytomas in paediatric patients.
Mossmann, D., Park, S. & Hall, M. N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757 (2018).
Molenaar, J. J. et al. Copy number defects of G1-cell cycle genes in neuroblastoma are frequent and correlate with high expression of E2F target genes and a poor prognosis. Genes Chromosomes Cancer 51, 10–19 (2012).
Geoerger, B. et al. A phase I study of the CDK4/6 inhibitor ribociclib (LEE011) in pediatric patients with malignant rhabdoid tumors, neuroblastoma, and other solid tumors. Clin. Cancer Res. 23, 2433–2441 (2017).
Ko, A., Han, S. Y. & Song, J. Regulatory network of ARF in cancer development. Mol. Cells 41, 381–389 (2018).
Phelps, D. et al. Inhibition of MDM2 by RG7388 confers hypersensitivity to X-radiation in xenograft models of childhood sarcoma. Pediatr. Blood Cancer 62, 1345–1352 (2015).
Van Goethem, A. et al. Dual targeting of MDM2 and BCL2 as a therapeutic strategy in neuroblastoma. Oncotarget 8, 57047–57057 (2017).
Howard, T. P. et al. MDM2 and MDM4 are therapeutic vulnerabilities in malignant rhabdoid tumors. Cancer Res. 79, 2404–2414 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03236857 (2019).
Bate-Eya, L. T. et al. High efficacy of the BCL-2 inhibitor ABT199 (venetoclax) in BCL-2 high-expressing neuroblastoma cell lines and xenografts and rational for combination with MCL-1 inhibition. Oncotarget 7, 27946–27958 (2016).
Robinson, G. W. et al. Vismodegib exerts targeted efficacy against recurrent sonic hedgehog-subgroup medulloblastoma: results from phase II Pediatric Brain Tumor Consortium studies PBTC-025B and PBTC-032. J. Clin. Oncol. 33, 2646–2654 (2015).
Robinson, G. W. et al. Irreversible growth plate fusions in children with medulloblastoma treated with a targeted hedgehog pathway inhibitor. Oncotarget 8, 69295–69302 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT01878617 (2019).
Durbin, A. D. et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat. Genet. 50, 1240–1246 (2018).
Chipumuro, E. et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139 (2014).
Puissant, A. et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 3, 308–323 (2013).
Witt, O., Deubzer, H. E., Lodrini, M., Milde, T. & Oehme, I. Targeting histone deacetylases in neuroblastoma. Curr. Pharm. Des. 15, 436–447 (2009).
Ecker, J. et al. Targeting class I histone deacetylase 2 in MYC amplified group 3 medulloblastoma. Acta Neuropathol. Commun. 3, 22 (2015).
Kim, K. H. et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 21, 1491–1496 (2015).
Kutko, M. C. et al. Histone deacetylase inhibitors induce growth suppression and cell death in human rhabdomyosarcoma in vitro. Clin. Cancer Res. 9, 5749–5755 (2003).
Hodges, T. R. et al. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro-oncology 19, 1047–1057 (2017).
Bouffet, E. et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 34, 2206–2211 (2016). This study reports durable responses of recurrent paediatric glioblastoma with biallelic mismatch repair deficiency to immune checkpoint inhibition.
Lord, C. J. & Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 16, 110–120 (2016).
Borkin, D. et al. Pharmacologic inhibition of the menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell 27, 589–602 (2015).
Garraway, L. A. et al. “Lineage addiction” in human cancer: lessons from integrated genomics. Cold Spring Harb. Symp. Quant. Biol. 70, 25–34 (2005).
Samstein, R. M. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206 (2019).
Havel, J. J., Chowell, D. & Chan, T. A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 19, 133–150 (2019).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Majzner, R. G., Heitzeneder, S. & Mackall, C. L. Harnessing the immunotherapy revolution for the treatment of childhood cancers. Cancer Cell 31, 476–485 (2017). This paper reviews immunotherapy options for the treatment of paediatric cancer, with a focus on immune checkpoint inhibitors and CAR T cells.
Groisberg, R. et al. Characteristics and outcomes of patients with advanced sarcoma enrolled in early phase immunotherapy trials. J. Immunother. Cancer 5, 100 (2017).
Lewin, J. et al. Response to immune checkpoint inhibition in two patients with alveolar soft-part sarcoma. Cancer Immunol. Res. 6, 1001–1007 (2018).
Davis, K. L. et al. ADVL1412: initial results of a phase I/II study of nivolumab and ipilimumab in pediatric patients with relapsed/refractory solid tumors — a COG study [abstract]. J. Clin. Oncol. 35 (Suppl. 15), 10526 (2017).
Geoerger, B. et al. Phase 1/2 KEYNOTE-051 study of pembrolizumab (pembro) in pediatric patients (pts) with advanced melanoma or a PD-L1+ advanced, relapsed, or refractory solid tumor or lymphoma [abstract]. J. Clin. Oncol. 35 (Suppl. 15), 10525 (2017).
Geoerger, B. et al. A phase I/II study of atezolizumab in pediatric and young adult patients with refractory/relapsed solid tumors (iMATRIX-Atezolizumab) [abstract]. J. Clin. Oncol. 35 (Suppl. 15), 10524 (2017).
Galluzzi, L., Buque, A., Kepp, O., Zitvogel, L. & Kroemer, G. Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell 28, 690–714 (2015).
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Greaves, M. F. Aetiology of acute leukaemia. Lancet 349, 344–349 (1997).
Johnson, K. J. et al. Childhood brain tumor epidemiology: a brain tumor epidemiology consortium review. Cancer Epidemiol. Biomarkers Prev. 23, 2716–2736 (2014).
Das, R. K., Vernau, L., Grupp, S. A. & Barrett, D. M. Naive T cell deficits at diagnosis and after chemotherapy impair cell therapy potential in pediatric cancers. Cancer Discov. 9, 492–499 (2019).
Griesinger, A. M. et al. Characterization of distinct immunophenotypes across pediatric brain tumor types. J. Immunol. 191, 4880–4888 (2013). This paper is one of the first to systematically characterize the frequency and phenotype of infiltrating immune cells in the most common paediatric brain tumour types.
Lin, G. L. et al. Non-inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol. Commun. 6, 51 (2018).
Plant, A. S. & Hwang, E. I. Immunotherapy and the immune infiltrate in pediatric brain tumors: an illustration and review of the unique challenges facing immunotherapy for pediatric oncology. Int. J. Immunol. Immunother. 5, 028 (2018).
Dhillon, S. Dinutuximab: first global approval. Drugs 75, 923–927 (2015).
Greenwood, K. & Foster, J. H. Dinutuximab for the treatment of pediatric patients with neuroblastoma. Drugs Today 53, 469–476 (2017).
Bosse, K. R. et al. Identification of GPC2 as an oncoprotein and candidate immunotherapeutic target in high-risk neuroblastoma. Cancer Cell 32, 295–309 (2017).
Mount, C. W. et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M+ diffuse midline gliomas. Nat. Med. 24, 572–579 (2018). This paper provides a promising and clinically available approach for future treatment of H3K27M-mutant diffuse midline gliomas, including DIPG, using CAR T cells.
Majzner, R. G. et al. CAR T cells targeting B7-H3, a pan-cancer antigen, demonstrate potent preclinical activity against pediatric solid tumors and brain tumors. Clin. Cancer Res. 25, 2560–2574 (2019).
Baldauf, M. C. et al. Systematic identification of cancer-specific MHC-binding peptides with RAVEN. Oncoimmunology 7, e1481558 (2018).
Pollack, I. F. et al. Antigen-specific immunoreactivity and clinical outcome following vaccination with glioma-associated antigen peptides in children with recurrent high-grade gliomas: results of a pilot study. J. Neurooncol. 130, 517–527 (2016).
Pollack, I. F. et al. Immune responses and outcome after vaccination with glioma-associated antigen peptides and poly-ICLC in a pilot study for pediatric recurrent low-grade gliomas. Neuro-oncology 18, 1157–1168 (2016).
van Gool, S. W. et al. Immunotherapy in atypical teratoid-rhabdoid tumors: data from a survey of the HGG-Immuno Group. Cytotherapy 18, 1178–1186 (2016).
Tsuchiya, N. et al. Phase I study of glypican-3-derived peptide vaccine therapy for patients with refractory pediatric solid tumors. Oncoimmunology 7, e1377872 (2017).
Merchant, M. S. et al. Adjuvant immunotherapy to improve outcome in high-risk pediatric sarcomas. Clin. Cancer Res. 22, 3182–3191 (2016).
[No authors listed]. Pembrolizumab approved for hodgkin lymphoma. Cancer Discov. 7, OF1 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02541604 (2019).
Merchant, M. S. et al. Phase I clinical trial of ipilimumab in pediatric patients with advanced solid tumors. Clin. Cancer Res. 22, 1364–1370 (2016).
Blumenthal, D. T. et al. Pembrolizumab: first experience with recurrent primary central nervous system (CNS) tumors. J. Neurooncol. 129, 453–460 (2016).
Pinto, N. et al. Patterns of PD-1, PD-L1, and PD-L2 expression in pediatric solid tumors. Pediatr. Blood Cancer 64, e26613 (2017).
Wedekind, M. F., Denton, N. L., Chen, C. Y. & Cripe, T. P. Pediatric cancer immunotherapy: opportunities and challenges. Paediatr. Drugs 20, 395–408 (2018).
Forrest, S. J., Geoerger, B. & Janeway, K. A. Precision medicine in pediatric oncology. Curr. Opin. Pediatr. 30, 17–24 (2018).
Tran, T. H., Shah, A. T. & Loh, M. L. Precision medicine in pediatric oncology: translating genomic discoveries into optimized therapies. Clin. Cancer Res. 23, 5329–5338 (2017).
DuBois, S. G., Corson, L. B., Stegmaier, K. & Janeway, K. A. Ushering in the next generation of precision trials for pediatric cancer. Science 363, 1175–1181 (2019). A comprehensive overview of different approaches to precision medicine clinical trial design and associated challenges.
Harris, M. H. et al. Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: the individualized cancer therapy (iCat) study. JAMA Oncol. 2, 608–615 (2016).
Harttrampf, A. C. et al. Molecular screening for cancer treatment optimization (MOSCATO-01) in pediatric patients: a single-institutional prospective molecular stratification trial. Clin. Cancer Res. 23, 6101–6112 (2017).
Mody, R. J. et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA 314, 913–925 (2015).
Oberg, J. A. et al. Implementation of next generation sequencing into pediatric hematology-oncology practice: moving beyond actionable alterations. Genome Med. 8, 133 (2016).
Parsons, D. W. et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2, 616–624 (2016).
Pincez, T. et al. Feasibility and clinical integration of molecular profiling for target identification in pediatric solid tumors. Pediatr. Blood Cancer 64, e26365 (2017).
Rusch, M. et al. Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat. Commun. 9, 3962 (2018).
Worst, B. C. et al. Next-generation personalised medicine for high-risk paediatric cancer patients – the INFORM pilot study. Eur. J. Cancer 65, 91–101 (2016).
Hill, R. M. et al. Combined MYC and p53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 27, 72–84 (2015).
Allen, C. E. et al. Target and agent prioritization for the Children’s Oncology Group-National Cancer Institute Pediatric MATCH Trial. J. Natl Cancer Inst. 109, djw274 (2017).
Capper, D. et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018). This paper describes a comprehensive molecular classification using DNA methylation patterns across a wide variety of human brain tumours.
Koelsche, C. et al. Array-based DNA-methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod. Pathol. 31, 1246–1256 (2018).
Renner, M. et al. Integrative DNA methylation and gene expression analysis in high-grade soft tissue sarcomas. Genome Biol. 14, r137 (2013).
Wu, S. P. et al. DNA methylation-based classifier for accurate molecular diagnosis of bone sarcomas. JCO Precis. Oncol. https://doi.org/10.1200/PO.17.00031 (2017).
Marron, J. M. et al. Patient/parent perspectives on genomic tumor profiling of pediatric solid tumors: the Individualized Cancer Therapy (iCat) experience. Pediatr. Blood Cancer 63, 1974–1982 (2016).
Mateo, J. et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann. Oncol. 29, 1895–1902 (2018).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03155620 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02813135 (2018).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02780128 (2019).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Waligora, M. et al. Risk and surrogate benefit for pediatric phase I trials in oncology: a systematic review with meta-analysis. PLOS Med. 15, e1002505 (2018).
Stewart, E. et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 549, 96–100 (2017).
Brabetz, S. et al. A biobank of patient-derived pediatric brain tumor models. Nat. Med. 24, 1752–1761 (2018).
Stewart, E. et al. The Childhood Solid Tumor Network: a new resource for the developmental biology and oncology research communities. Dev. Biol. 411, 287–293 (2016).
Meehan, T. F. et al. PDX-MI: minimal information for patient-derived tumor xenograft models. Cancer Res. 77, e62–e66 (2017).
Rees, M. G. et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 12, 109–116 (2016).
Seashore-Ludlow, B. et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 5, 1210–1223 (2015).
Yang, W. et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2013).
Dhanjal, J. K., Radhakrishnan, N. & Sundar, D. Identifying synthetic lethal targets using CRISPR/Cas9 system. Methods 131, 66–73 (2017).
Smith, M. A. & Houghton, P. A proposal regarding reporting of in vitro testing results. Clin. Cancer Res. 19, 2828–2833 (2013).
Murphy, B. et al. Evaluation of alternative in vivo drug screening methodology: a single mouse analysis. Cancer Res. 76, 5798–5809 (2016).
Yao, Y. M. et al. Mouse PDX trial suggests synergy of concurrent inhibition of RAF and EGFR in colorectal cancer with BRAF or KRAS mutations. Clin. Cancer Res. 23, 5547–5560 (2017).
Gao, H. et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 21, 1318–1325 (2015). This study describes the use of the ‘single mouse’ design for high-throughput in vivo screening of agents for biomarker discovery using genomically characterized PDX models.
Nimmervoll, B. V. et al. Establishing a preclinical multidisciplinary board for brain tumors. Clin. Cancer Res. 24, 1654–1666 (2018).
Rose, W. C. & Wild, R. Therapeutic synergy of oral taxane BMS-275183 and cetuximab versus human tumor xenografts. Clin. Cancer Res. 10, 7413–7417 (2004).
Houghton, P. J. et al. Stage 2 combination testing of rapamycin with cytotoxic agents by the Pediatric Preclinical Testing Program. Mol. Cancer Ther. 9, 101–112 (2010). This study describes a systematic approach to in vivo combination testing using paediatric preclinical models.
Komlodi-Pasztor, E., Sackett, D. L. & Fojo, A. T. Inhibitors targeting mitosis: tales of how great drugs against a promising target were brought down by a flawed rationale. Clin. Cancer Res. 18, 51–63 (2012).
Furman, W. L. et al. Direct translation of a protracted irinotecan schedule from a xenograft model to a phase I trial in children. J. Clin. Oncol. 17, 1815–1824 (1999).
Jacus, M. O. et al. Deriving therapies for children with primary CNS tumors using pharmacokinetic modeling and simulation of cerebral microdialysis data. Eur. J. Pharm. Sci. 57, 41–47 (2014).
Carol, H. et al. The anti-CD19 antibody-drug conjugate SAR3419 prevents hematolymphoid relapse postinduction therapy in preclinical models of pediatric acute lymphoblastic leukemia. Clin. Cancer Res. 19, 1795–1805 (2013).
Sano, R. et al. Pediatric Preclinical Testing Consortium evaluation of a DLL3-targeted antibody drug conjugate rovalpituzumab tesirine, in neuroblastoma [abstract]. Cancer Res. 78 (Suppl. 13), LB-136 (2018).
Bacac, M. et al. CD20-TCB with obinutuzumab pretreatment as next-generation treatment of hematologic malignancies. Clin. Cancer Res. 24, 4785–4797 (2018). This paper describes methods for in vivo testing of T cell-engaging immuno-oncology agents that can be applied to paediatric preclinical testing.
Drilon, A. et al. Efficacy of larotrectinib in TRK fusion–positive cancers in adults and children. N. Engl. J. Med. 378, 731–739 (2018).
Yang, W. et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat. Med. 25, 767–775 (2019).
Lin, C. Y. et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530, 57–62 (2016).
Mack, S. C. et al. Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature 553, 101 (2017).
Pearson, A. D. J. et al. From class waivers to precision medicine in paediatric oncology. Lancet Oncol. 18, e394–e404 (2017).
Moreno, L. et al. Early phase clinical trials of anticancer agents in children and adolescents — an ITCC perspective. Nat. Rev. Clin. Oncol. 14, 497 (2017).
Acknowledgements
The authors acknowledge the valuable contribution from E. Mould and V. Tyrrell to the design of figure 2 and manuscript content. We also thank all those contributing to the personalized medicine programmes listed as well as the patients and their families, who have made the advances outlined here possible by consenting to take part in scientific and/or clinical studies.
Author information
Authors and Affiliations
Contributions
D.T.W.J., A.B., T.G.P.G., M.H., N.J., M.K., T.M., J.J.M., A.N., T.J.P., G.S., M.A.S., F.W. and S.M.P. researched the content for the article, discussed and prepared the text and display items, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Cancer thanks N. Gottardo, K. Stegmaier and M. Roussel for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
ACCELERATE: www.accelerate-platform.eu
Cancerrxgene: Genomics of Drug Sensitivity in Cancer: www.cancerrxgene.org
Cavatica: www.cavatica.org
ClinicalTrials.gov: https://clinicaltrials.gov/
CONNECT: www.connectconsortium.org
Depmap: the Cancer Dependency map: http://depmap.org
ITCC: www.itcc-consortium.org
ITCC-P4: www.itccp4.eu
PBTC: www.pbtc.org
PeCan: http://pecan.stjude.cloud
PedcBioPortal: http://pedcbioportal.org
PNOC: www.pnoc.us
PPTC: www.ncipptc.org
R2: http://r2.amc.nl
The Treehouse Childhood Cancer Initiative: https://treehousegenomics.soe.ucsc.edu/
Glossary
- Radial glia cells
-
Ventricular zone progenitor cells characterized by long radial processes that support neuron formation and migration.
- Supratentorial
-
Referring to a brain region located in the area above the tentorium cerebelli that contains the cerebral hemispheres along with the basal ganglia, thalamus and other midline structures.
- Posterior fossa
-
Also known as the infratentorial region. A brain region located in the area below the tentorium cerebelli that contains the cerebellum and brain stem.
- Embryonal tumours
-
Tumours originating from embryonic (fetal) tissue.
- SWI/SNF complex
-
Multiprotein complex responsible for nucleosome remodelling.
- Uniparental disomy
-
A type of copy-neutral structural variation, characterized as the same-parent origin of both chromosomes of a homologous chromosome pair.
- Telomere maintenance mechanisms
-
(TMMs). To avoid critical telomere shortening due to successive cell divisions, tumour cells can adopt telomere maintenance mechanisms either by reactivation of the telomerase enzyme or by the telomerase-independent, recombination-mediated alternative lengthening of telomeres (ALT) pathway.
- Risk-adapted therapy
-
Strategy consisting of specific treatment protocols based on individual risk stratification and designed to increase the likelihood of cure while minimizing late therapy-related effects.
- Pilocytic astrocytoma
-
A World Health Organization (WHO) Grade I tumour thought to be of astrocytic origin, primarily arising during childhood and typically driven by alterations of the MAPK signalling pathway. ‘Pilocytic’ refers to the elongated, hair-like (piloid) processes that often characterize the tumour cells.
- Nutlin
-
A family of small molecules identified from a high-throughput screen that inhibit the interaction between MDM2 and p53, with the aim of re-activating p53 function in cells where wild-type p53 is inhibited by elevated MDM2 levels.
- Replication stress
-
A process occurring typically during DNA replication when the genome is exposed to one of a number of stressful stimuli, which can result in stalled replication forks and other errors in the cell cycle.
- Neurological sequelae
-
Long-term complications of central nervous system disease, which can affect cognitive, sensory and motor functions, thereby negatively impacting the quality of life of a patient long after the original disease process is cured.
- Antibody–drug conjugate
-
Complex molecule composed of a monoclonal antibody linked to a biologically active cytotoxic anticancer agent.
- Pharmacogenomics
-
A combination of pharmacology and genomics, this relatively new field investigates how variants in genes involved in different aspects of drug processing can affect an individual’s response to a given therapy.
- Pharmacokinetic profile
-
A description of how the processes of absorption and distribution, as well as metabolic processing and excretion, affect the levels of a drug present within the body in the period after its administration.
- T cell bispecific antibodies
-
Antibodies that can simultaneously bind to CD3ε, a component of the T cell receptor complex, and to a tumour antigen, enabling the formation of an immunological synapse between the tumour cell and the T cell that results in tumour killing.
Rights and permissions
About this article
Cite this article
Jones, D.T.W., Banito, A., Grünewald, T.G.P. et al. Molecular characteristics and therapeutic vulnerabilities across paediatric solid tumours. Nat Rev Cancer 19, 420–438 (2019). https://doi.org/10.1038/s41568-019-0169-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-019-0169-x
This article is cited by
-
CNS tumor stroma transcriptomics identify perivascular fibroblasts as predictors of immunotherapy resistance in glioblastoma patients
npj Genomic Medicine (2023)
-
Molecular genomic landscape of pediatric solid tumors in Chinese patients: implications for clinical significance
Journal of Cancer Research and Clinical Oncology (2023)
-
PatchResNet: Multiple Patch Division–Based Deep Feature Fusion Framework for Brain Tumor Classification Using MRI Images
Journal of Digital Imaging (2023)
-
Transcriptional immunogenomic analysis reveals distinct immunological clusters in paediatric nervous system tumours
Genome Medicine (2023)
-
Real-world performance analysis of a novel computational method in the precision oncology of pediatric tumors
World Journal of Pediatrics (2023)