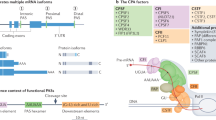
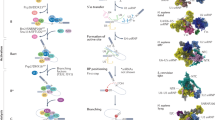
Abstract
Most human genes have multiple sites at which RNA 3ʹ end cleavage and polyadenylation can occur, enabling the expression of distinct transcript isoforms under different conditions. Novel methods to sequence RNA 3′ ends have generated comprehensive catalogues of polyadenylation (poly(A)) sites; their analysis using innovative computational methods has revealed how poly(A) site choice is regulated by core RNA 3ʹ end processing factors, such as cleavage factor I and cleavage and polyadenylation specificity factor, as well as by other RNA-binding proteins, particularly splicing factors. Here, we review the experimental and computational methods that have enabled the global mapping of mRNA and of long non-coding RNA 3ʹ ends, quantification of the resulting isoforms and the discovery of regulators of alternative cleavage and polyadenylation (APA). We highlight the different types of APA-derived isoforms and their functional differences, and illustrate how APA contributes to human diseases, including cancer and haematological, immunological and neurological diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Reyes, A. & Huber, W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res. 46, 582–592 (2018).
Edmonds, M., Vaughan, M. H. Jr & Nakazato, H. Polyadenylic acid sequences in the heterogeneous nuclear RNA and rapidly-labeled polyribosomal RNA of HeLa cells: possible evidence for a precursor relationship. Proc. Natl Acad. Sci. USA 68, 1336–1340 (1971).
Darnell, J. E., Wall, R. & Tushinski, R. J. An adenylic acid-rich sequence in messenger RNA of HeLa cells and its possible relationship to reiterated sites in DNA. Proc. Natl Acad. Sci. USA 68, 1321–1325 (1971).
Tian, B., Hu, J., Zhang, H. & Lutz, C. S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212 (2005).
Mayr, C. Evolution and biological roles of alternative 3ʹUTRs. Trends Cell Biol. 26, 227–237 (2016).
Lianoglou, S., Garg, V., Yang, J. L., Leslie, C. S. & Mayr, C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396 (2013).
Martin, G., Gruber, A. R., Keller, W. & Zavolan, M. Genome-wide analysis of pre-mRNA 3ʹ end processing reveals a decisive role of human cleavage factor I in the regulation of 3ʹ UTR length. Cell Rep. 1, 753–763 (2012).
Li, W. et al. Systematic profiling of poly (A)+ transcripts modulated by core 3ʹ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLOS Genet. 11, e1005166 (2015).
Brumbaugh, J. et al. Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell 172, 106–120 (2018). This paper demonstrates that components of the 3ʹ end processing complex influence cell fate, presumably through the APA of chromatin factors.
Xia, Z. et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3ʹ-UTR landscape across seven tumour types. Nat. Commun. 5, 5274 (2014). Here, the analysis of 3ʹ UTR usage in tumour cells compared to normal cells reveals the general shortening of 3ʹ UTRs in cancers.
Gruber, A. J. et al. Discovery of physiological and cancer-related regulators of 3ʹ UTR processing with KAPAC. Genome Biol. 19, 44 (2018). This study describes the PAQR tool for quantifying poly(A) site usage from RNA-seq data and the KAPAC tool for discovering regulatory motifs that influence polyadenylation based on the quantification of poly(A) site usage.
Leung, M. K. K., Delong, A. & Frey, B. J. Inference of the human polyadenylation code. Bioinformatics 34, 2889–2898 (2018).
Shi, Y. et al. Molecular architecture of the human pre-mRNA 3ʹ processing complex. Mol. Cell 33, 365–376 (2009).
Tian, B. & Manley, J. L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell. Biol. 18, 18–30 (2017).
Zhao, J., Hyman, L. & Moore, C. Formation of mRNA 3ʹ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445 (1999).
Hill, C. H. et al. Activation of the endonuclease that defines mRNA 3ʹ ends requires incorporation into an 8-subunit core cleavage and polyadenylation factor complex. Mol. Cell 73, 1217–1231 (2019).
Proudfoot, N. J. & Brownlee, G. G. 3ʹ non-coding region sequences in eukaryotic messenger RNA. Nature 263, 211–214 (1976).
Gruber, A. J. et al. A comprehensive analysis of 3ʹ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 26, 1145–1159 (2016).
Dantonel, J. C., Murthy, K. G., Manley, J. L. & Tora, L. Transcription factor TFIID recruits factor CPSF for formation of 3ʹ end of mRNA. Nature 389, 399–402 (1997).
Murthy, K. G. & Manley, J. L. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J. Biol. Chem. 267, 14804–14811 (1992).
Kaufmann, I., Martin, G., Friedlein, A., Langen, H. & Keller, W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23, 616–626 (2004).
Keller, W., Bienroth, S., Lang, K. M. & Christofori, G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3ʹ processing signal AAUAAA. EMBO J. 10, 4241–4249 (1991).
Schönemann, L. et al. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 28, 2381–2393 (2014).
Chan, S. L. et al. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3ʹ processing. Genes Dev. 28, 2370–2380 (2014).
Clerici, M., Faini, M., Aebersold, R. & Jinek, M. Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex. eLife 6, e33111 (2017).
Sun, Y. et al. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc. Natl Acad. Sci. USA 115, E1419–E1428 (2018).
Beaudoing, E., Freier, S., Wyatt, J. R., Claverie, J. M. & Gautheret, D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10, 1001–1010 (2000).
Sheets, M. D., Ogg, S. C. & Wickens, M. P. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 18, 5799–5805 (1990). This paper provides an in vitro estimation of the efficiency of variants of AAUAAA on cleavage and polyadenylation.
Gruber, A. R., Martin, G., Keller, W. & Zavolan, M. Means to an end: mechanisms of alternative polyadenylation of messenger RNA precursors. Wiley Interdiscip. Rev. RNA 5, 183–196 (2013).
Wang, R., Zheng, D., Yehia, G. & Tian, B. A compendium of conserved cleavage and polyadenylation events in mammalian genes. Genome Res. 28, 1427–1441 (2018).
Dominski, Z., Yang, X.-C. & Marzluff, W. F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123, 37–48 (2005).
Mandel, C. R. et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3ʹ-end-processing endonuclease. Nature 444, 953–956 (2006).
Kühn, U. et al. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284, 22803–22814 (2009).
Eckmann, C. R., Rammelt, C. & Wahle, E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA 2, 348–361 (2011).
Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H. & Bartel, D. P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014).
Lim, J., Lee, M., Son, A., Chang, H. & Kim, V. N. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev. 30, 1671–1682 (2016).
Park, J.-E., Yi, H., Kim, Y., Chang, H. & Kim, V. N. Regulation of poly(A) tail and translation during the somatic cell cycle. Mol. Cell 62, 462–471 (2016).
Lima, S. A. et al. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 24, 1057–1063 (2017).
MacDonald, C. C., Wilusz, J. & Shenk, T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 14, 6647–6654 (1994).
Takagaki, Y., Seipelt, R. L., Peterson, M. L. & Manley, J. L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87, 941–952 (1996).
Wallace, A. M. et al. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc. Natl Acad. Sci. USA 96, 6763–6768 (1999).
Yao, C. et al. Overlapping and distinct functions of CstF64 and CstF64τ in mammalian mRNA 3ʹ processing. RNA 19, 1781–1790 (2013).
Yao, C. et al. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc. Natl Acad. Sci. USA 109, 18773–18778 (2012).
Venkataraman, K., Brown, K. M. & Gilmartin, G. M. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 19, 1315–1327 (2005).
Yang, Q., Gilmartin, G. M. & Doublié, S. Structural basis of UGUA recognition by the Nudix protein CFIm25 and implications for a regulatory role in mRNA 3ʹ processing. Proc. Natl Acad. Sci. USA 107, 10062–10067 (2010).
Yang, Q., Coseno, M., Gilmartin, G. M. & Doublié, S. Crystal structure of a human cleavage factor CFIm25/CFIm68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure 19, 368–377 (2011).
Schäfer, P. et al. Reconstitution of mammalian cleavage factor II involved in 3ʹ processing of mRNA precursors. RNA 24, 1721–1737 (2018).
Derti, A. et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 22, 1173–1183 (2012).
Wang, R., Nambiar, R., Zheng, D. & Tian, B. PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res. 46, D315–D319 (2018).
You, L. et al. APASdb: a database describing alternative poly (A) sites and selection of heterogeneous cleavage sites downstream of poly (A) signals. Nucleic Acids Res. 43, D59–D67 (2014).
Gruber, A. R. et al. Global 3ʹ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat. Commun. 5, 5465 (2014).
Kanitz, A. et al. Comparative assessment of methods for the computational inference of transcript isoform abundance from RNA-seq data. Genome Biol. 16, 150 (2015).
Rot, G. et al. High-resolution RNA maps suggest common principles of splicing and polyadenylation regulation by TDP-43. Cell Rep. 19, 1056–1067 (2017).
Wang, W., Wei, Z. & Li, H. A change-point model for identifying 3ʹ UTR switching by next-generation RNA sequencing. Bioinformatics 30, 2162–2170 (2014).
Weinstein, J. N. et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Ha, K. C. H., Blencowe, B. J. & Morris, Q. QAPA: a new method for the systematic analysis of alternative polyadenylation from RNA-seq data. Genome Biol. 19, 45 (2018).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Ye, C., Long, Y., Ji, G., Li, Q. Q. & Wu, X. APAtrap: identification and quantification of alternative polyadenylation sites from RNA-seq data. Bioinformatics 34, 1841–1849 (2018).
Chang, J.-W. et al. An integrative model for alternative polyadenylation, IntMAP, delineates mTOR-modulated endoplasmic reticulum stress response. Nucleic Acids Res. 46, 5996–6008 (2018).
Hwang, H.-W. et al. PAPERCLIP identifies microRNA targets and a role of CstF64/64tau in promoting non-canonical poly(A) site usage. Cell Rep. 15, 423–435 (2016).
Elkon, R., Ugalde, A. P. & Agami, R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 14, 496–506 (2013).
Mayr, C. What are 3ʹ UTRs doing? Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a034728 (2018).
Berkovits, B. D. & Mayr, C. Alternative 3ʹ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367 (2015). This study uncovers the mechanism of 3ʹ UTR-dependent protein localization.
Ciolli Mattioli, C. et al. Alternative 3ʹ UTRs direct localization of functionally diverse protein isoforms in neuronal compartments. Nucleic Acids Res. 47, 2560–2573 (2019).
Sandberg, R., Neilson, J. R., Sarma, A., Sharp, P. A. & Burge, C. B. Proliferating cells express mRNAs with shortened 3ʹ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647 (2008). This study demonstrates that proliferating cells express transcripts with shorter 3ʹ UTRs than non-proliferating cells.
Mayr, C. & Bartel, D. P. Widespread shortening of 3ʹUTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 (2009).
Ji, Z. & Tian, B. Reprogramming of 3ʹ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLOS ONE 4, e8419 (2009).
Elkon, R. et al. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 13, R59 (2012).
Lackford, B. et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 33, 878–889 (2014).
Berg, M. G. et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150, 53–64 (2012).
Fanourgakis, G., Lesche, M., Akpinar, M., Dahl, A. & Jessberger, R. Chromatoid body protein TDRD6 supports long 3ʹ UTR triggered nonsense mediated mRNA decay. PLOS Genet. 12, e1005857 (2016).
Bao, J. et al. UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3ʹUTR transcripts. PLOS Genet. 12, e1005863 (2016).
Neve, J., Patel, R., Wang, Z., Louey, A. & Furger, A. M. Cleavage and polyadenylation: ending the message expands gene regulation. RNA Biol. 14, 865–890 (2017).
Crawford, E. K., Ensor, J. E., Kalvakolanu, I. & Hasday, J. D. The role of 3ʹ poly(A) tail metabolism in tumor necrosis factor-α regulation. J. Biol. Chem. 272, 21120–21127 (1997).
Peattie, D. A., Hsiao, K., Benasutti, M. & Lippke, J. A. Three distinct messenger RNAs can encode the human immunosuppressant-binding protein FKBP12. Gene 150, 251–257 (1994).
Chuvpilo, S. et al. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity 10, 261–269 (1999).
Alt, F. W. et al. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3ʹ ends. Cell 20, 293–301 (1980).
Singh, I. et al. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nat. Commun. 9, 1716 (2018).
Gruber, A. J., Gypas, F., Riba, A., Schmidt, R. & Zavolan, M. Terminal exon characterization with TECtool reveals an abundance of cell-specific isoforms. Nat. Methods 15, 832–836 (2018). In this study, the use of TECtool identifies terminal exons ending at ‘intronic’ poly(A) sites and shows their high prevalence in immune cells and in germ cells.
Spies, N., Burge, C. B. & Bartel, D. P. 3ʹ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 23, 2078–2090 (2013).
Gaidatzis, D., van Nimwegen, E., Hausser, J. & Zavolan, M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics 8, 69 (2007).
Hoffman, Y. et al. 3ʹUTR shortening potentiates microRNA-based repression of pro-differentiation genes in proliferating human cells. PLOS Genet. 12, e1005879 (2016).
Plass, M., Rasmussen, S. H. & Krogh, A. Highly accessible AU-rich regions in 3ʹ untranslated regions are hotspots for binding of regulatory factors. PLOS Comput. Biol. 13, e1005460 (2017).
Zarnack, K. et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152, 453–466 (2013).
Ma, W. & Mayr, C. A. Membraneless organelle associated with the endoplasmic reticulum enables 3ʹ UTR-mediated protein-protein interactions. Cell 175, 1492–1506 (2018). The paper reports the identification of TIS RNA granules and of the TIGER compartment, which form through 3´ UTR-mediated interactions with TIS11B.
Goers, E. S., Purcell, J., Voelker, R. B., Gates, D. P. & Berglund, J. A. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 38, 2467–2484 (2010).
Taliaferro, J. M. et al. Distal alternative last exons localize mRNAs to neural projections. Mol. Cell 61, 821–833 (2016). The study demonstrates that 3ʹ UTRs play a general role in transcript localization in neurons.
Yudin, D. et al. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron 59, 241–252 (2008).
An, J. J. et al. Distinct role of long 3ʹ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134, 175–187 (2008).
Gruber, A. R., Martin, G., Keller, W. & Zavolan, M. Cleavage factor Im is a key regulator of 3ʹ UTR length. RNA Biol. 9, 1405–1412 (2012).
Zhu, Y. et al. Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol. Cell 69, 62–74 (2018).
Luo, W. et al. The conserved intronic cleavage and polyadenylation site of CstF-77 gene imparts control of 3ʹ end processing activity through feedback autoregulation and by U1 snRNP. PLOS Genet. 9, e1003613 (2013).
Niwa, M., Rose, S. D. & Berget, S. M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4, 1552–1559 (1990).
Kyburz, A., Friedlein, A., Langen, H. & Keller, W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3ʹ end processing and splicing. Mol. Cell 23, 195–205 (2006).
Millevoi, S. et al. An interaction between U2AF 65 and CF Im links the splicing and 3ʹ end processing machineries. EMBO J. 25, 4854–4864 (2006).
Almada, A. E., Wu, X., Kriz, A. J., Burge, C. B. & Sharp, P. A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363 (2013).
Wahl, M. C., Will, C. L. & Lührmann, R. The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 (2009).
Kaida, D. et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468, 664–668 (2010). This study demonstrates that the U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation.
Oh, J.-M. et al. U1 snRNP telescripting regulates a size-function-stratified human genome. Nat. Struct. Mol. Biol. 24, 993–999 (2017).
Ule, J. et al. An RNA map predicting Nova-dependent splicing regulation. Nature 444, 580–586 (2006).
Singh, R., Valcárcel, J. & Green, M. R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268, 1173–1176 (1995).
Polydorides, A. D., Okano, H. J., Yang, Y. Y., Stefani, G. & Darnell, R. B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl Acad. Sci. USA 97, 6350–6355 (2000).
Lou, H., Gagel, R. F. & Berget, S. M. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 10, 208–219 (1996).
Zhu, H., Zhou, H.-L., Hasman, R. A. & Lou, H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 282, 2203–2210 (2007).
Dai, W., Zhang, G. & Makeyev, E. V. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res. 40, 787–800 (2012).
Izquierdo, J. M. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 283, 19077–19084 (2008).
Tajnik, M. et al. Intergenic Alu exonisation facilitates the evolution of tissue-specific transcript ends. Nucleic Acids Res. 43, 10492–10505 (2015).
Ji, X. et al. αCP binding to a cytosine-rich subset of polypyrimidine tracts drives a novel pathway of cassette exon splicing in the mammalian transcriptome. Nucleic Acids Res. 44, 2283–2297 (2016).
Ji, X., Wan, J., Vishnu, M., Xing, Y. & Liebhaber, S. A. αCP Poly(C) binding proteins act as global regulators of alternative polyadenylation. Mol. Cell. Biol. 33, 2560–2573 (2013).
Makeyev, A. V. & Liebhaber, S. A. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8, 265–278 (2002).
Jenal, M. et al. The poly (A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538–553 (2012).
Hosoda, N., Lejeune, F. & Maquat, L. E. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 26, 3085–3097 (2006).
Zheng, D. & Tian, B. in Systems Biology of RNA Binding Proteins (ed. Yeo, G. W.) 97–127 (Springer, NY, 2014).
Masuda, A. et al. Position-specific binding of FUS to nascent RNA regulates mRNA length. Genes Dev. 29, 1045–1057 (2015).
Schwartz, J. C., Cech, T. R. & Parker, R. R. Biochemical properties and biological functions of FET proteins. Annu. Rev. Biochem. 84, 355–379 (2015).
Giudice, G., Sánchez-Cabo, F., Torroja, C. & Lara-Pezzi, E. ATtRACT-a database of RNA-binding proteins and associated motifs. Database 2016, baw035 (2016).
Curinha, A., Oliveira Braz, S., Pereira-Castro, I., Cruz, A. & Moreira, A. Implications of polyadenylation in health and disease. Nucleus 5, 508–519 (2014).
Chang, J. W., Yeh, H. S. & Yong, J. Alternative polyadenylation in human diseases. Endocrinol. Metab. 32, 413–421 (2017).
Bacchetta, R., Barzaghi, F. & Roncarolo, M.-G. From IPEX syndrome to FOXP3 mutation: a lesson on immune dysregulation. Ann. NY Acad. Sci. 1417, 5–22 (2016).
Bennett, C. L. et al. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA→AAUGAA) leads to the IPEX syndrome. Immunogenetics 53, 435–439 (2001).
Stacey, S. N. et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat. Genet. 43, 1098–1103 (2011).
Garin, I. et al. Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. Proc. Natl Acad. Sci. USA 107, 3105–3110 (2010).
Higgs, D. R. et al. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature 306, 398–400 (1983).
Orkin, S. H., Cheng, T. C., Antonarakis, S. E. & Kazazian, H. H. Jr. Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J. 4, 453–456 (1985).
Hellquist, A. et al. The human GIMAP5 gene has a common polyadenylation polymorphism increasing risk to systemic lupus erythematosus. J. Med. Genet. 44, 314–321 (2007).
Graham, R. R. et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl Acad. Sci. USA 104, 6758–6763 (2007).
Lane, D. A. & Grant, P. J. Role of hemostatic gene polymorphisms in venous and arterial thrombotic disease. Blood 95, 1517–1532 (2000).
Ferraresi, P. et al. The heterozygous 20210 G/A prothrombin genotype is associated with early venous thrombosis in inherited thrombophilias and is not increased in frequency in artery disease. Arterioscler. Thromb. Vasc. Biol. 17, 2418–2422 (1997).
Ridker, P. M., Hennekens, C. H. & Miletich, J. P. G20210A mutation in prothrombin gene and risk of myocardial infarction, stroke, and venous thrombosis in a large cohort of US men. Circulation 99, 999–1004 (1999).
Gehring, N. H. et al. Increased efficiency of mRNA 3ʹ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat. Genet. 28, 389–392 (2001).
Ceelie, H., Spaargaren-van Riel, C. C., Bertina, R. M. & Vos, H. L. G20210A is a functional mutation in the prothrombin gene; effect on protein levels and 3ʹ-end formation. J. Thromb. Haemost. 2, 119–127 (2004).
Poort, S. R., Rosendaal, F. R., Reitsma, P. H. & Bertina, R. M. A common genetic variation in the 3ʹ-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood 88, 3698–3703 (1996).
Brais, B. et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 18, 164–167 (1998).
Singh, P. et al. Global changes in processing of mRNA 3ʹ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 69, 9422–9430 (2009).
Chang, J.-W. et al. mRNA 3ʹ-UTR shortening is a molecular signature of mTORC1 activation. Nat. Commun. 6, 7218 (2015).
Xue, Z. et al. Recurrent tumor-specific regulation of alternative polyadenylation of cancer-related genes. BMC Genomics 19, 536 (2018).
Masamha, C. P. et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510, 412–416 (2014).
Lee, S.-H. et al. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature 561, 127–131 (2018). The study reveals that intronic polyadenylation is widespread in leukaemia and that it leads to inactivation of tumor suppressors.
Wiestner, A. et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood 109, 4599–4606 (2007).
Decorsière, A. et al. Decreased efficiency of MSH6 mRNA polyadenylation linked to a 20-base-pair duplication in Lynch syndrome families. Cell Cycle 11, 2578–2580 (2012).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Van Nostrand, E. L. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016).
Floor, S. N. & Doudna, J. A. Tunable protein synthesis by transcript isoforms in human cells. eLife 5, e10921 (2016).
Coulon, A. et al. Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife 3, e03939 (2014).
Vorlová, S. et al. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol. Cell 43, 927–939 (2011).
Van Etten, J. L. et al. Targeting a single alternative polyadenylation site coordinately blocks expression of androgen receptor mRNA splice variants in prostate cancer. Cancer Res. 77, 5228–5235 (2017).
Yoon, O. K. & Brem, R. B. Noncanonical transcript forms in yeast and their regulation during environmental stress. RNA 16, 1256–1267 (2010).
Ozsolak, F. et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143, 1018–1029 (2010).
Fox-Walsh, K., Davis-Turak, J., Zhou, Y., Li, H. & Fu, X.-D. A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3ʹ end formation. Genomics 98, 266–271 (2011).
Shepard, P. J. et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17, 761–772 (2011).
Jan, C. H., Friedman, R. C., Ruby, J. G. & Bartel, D. P. Formation, regulation and evolution of Caenorhabditis elegans 3ʹUTRs. Nature 469, 97–101 (2011).
Fu, Y. et al. Differential genome-wide profiling of tandem 3ʹ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 21, 741–747 (2011).
Hoque, M. et al. Analysis of alternative cleavage and polyadenylation by 3ʹ region extraction and deep sequencing. Nat. Methods 10, 133–139 (2013).
Pelechano, V., Wei, W., Jakob, P. & Steinmetz, L. M. Genome-wide identification of transcript start and end sites by transcript isoform sequencing. Nat. Protoc. 9, 1740–1759 (2014).
Sanfilippo, P., Miura, P. & Lai, E. C. Genome-wide profiling of the 3ʹ ends of polyadenylated RNAs. Methods 126, 86–94 (2017).
Routh, A. et al. Poly(A)-ClickSeq: click-chemistry for next-generation 3ʹ-end sequencing without RNA enrichment or fragmentation. Nucleic Acids Res. 45, e112 (2017).
Cook, K. B., Kazan, H., Zuberi, K., Morris, Q. & Hughes, T. R. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 39, D301–D308 (2011).
Grassi, E., Mariella, E., Lembo, A., Molineris, I. & Provero, P. Roar: detecting alternative polyadenylation with standard mRNA sequencing libraries. BMC Bioinformatics 17, 423 (2016).
Arefeen, A., Liu, J., Xiao, X. & Jiang, T. TAPAS: tool for alternative polyadenylation site analysis. Bioinformatics 34, 2521–2529 (2018).
Harteveld, C. L. et al. A novel polyadenylation signal mutation in the alpha 2-globin gene causing alpha thalassaemia. Br. J. Haematol. 87, 139–143 (1994).
Rund, D. et al. Two mutations in the beta-globin polyadenylylation signal reveal extended transcripts and new RNA polyadenylylation sites. Proc. Natl Acad. Sci. USA 89, 4324–4328 (1992).
Andreu, N. et al. A novel Wiskott-Aldrich syndrome protein (WASP) complex mutation identified in a WAS patient results in an aberrant product at the C-terminus from two transcripts with unusual polyA signals. J. Hum. Genet. 51, 92–97 (2006).
Shin, J.-H. et al. IA-2 autoantibodies in incident type I diabetes patients are associated with a polyadenylation signal polymorphism in GIMAP5. Genes Immun. 8, 503–512 (2007).
Acknowledgements
The authors thank G. Martin and W. Keller for numerous discussions.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
ENSEMBL: https://www.ensembl.org/
STRINGTIE: https://ccb.jhu.edu/software/stringtie/
The Cancer Genome Atlas: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga
Glossary
- Poly(A) sites
-
(Also referred to as CPA sites or 3ʹ end processing sites). Single nucleotides after which a pre-mRNA is cleaved and the non-template synthesis of the poly(A) tail is initiated.
- Alternative cleavage and polyadenylation
-
(APA). The process by which the distinct poly(A) sites of a gene are processed under distinct conditions to give rise to different transcript isoforms.
- 3′ UTR isoforms
-
Isoforms that differ only in their 3′ untranslated regions (UTRs). They are generated through RNA 3′ end processing at alternative poly(A) sites.
- Poly(A) signal
-
(PAS). The main sequence element that defines 3′ end processing sites. It is bound by the WDR33 and CPSF4 proteins of the CPSF complex and most commonly has the sequence AAUAAA.
- Terminal exon
-
The last exon of a transcript.
- Cassette TEs
-
Exons that are included as terminal exons in some transcripts generated from a gene but spliced out in others.
- Effector T cells
-
Relatively short-lived T lymphocytes that have been ‘primed’ by various signals and are ready to interact with the pathogen.
- Small nuclear ribonucleoprotein
-
(snRNP). An RNA–protein complex that participates in RNA splicing, together with proteins and pre-mRNAs. The snRNPs are U1, U2, U4, U5, U6, U11, U12 and U4atac.
- Adaptive immune response
-
The defence mechanism against microbial pathogens in vertebrates. It is based on a complex immune system that is trained during development to distinguish structures germane to the organism (self) from external structures (non-self).
- Cis-regulatory elements
-
Regions in a nucleic acid that regulate the expression of the associated gene and transcript.
- Telescripting
-
The small nuclear ribonucleoprotein U1 suppresses premature cleavage and polyadenylation at cryptic poly(A) sites in nascent RNA polymerase II transcripts. As this suppressive activity of U1 is required for full-length gene transcription, it is termed ‘telescripting’.
- Alu transposable elements
-
Short, mobile DNA elements that can change (transpose) their position in the genome with the help of trans-acting factors.
- Crosslinking and immunoprecipitation
-
A method for identifying the targets of RNA-binding proteins (RBPs). It involves crosslinking a protein of interest to RNAs using ultraviolet irradiation, followed by partial RNA digestion, immunoprecipitation of the protein with bound RNAs and sequencing of RBP-bound RNA fragments.
- Single-nucleotide polymorphisms
-
(SNPs). Variations at a defined position in the genome; some individuals in the population carry a particular nucleotide at that position, while other individuals carry a different nucleotide.
Rights and permissions
About this article
Cite this article
Gruber, A.J., Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet 20, 599–614 (2019). https://doi.org/10.1038/s41576-019-0145-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-019-0145-z
This article is cited by
-
Genome-wide characterization of post-transcriptional processes related to wood formation in Dalbergia odorifera
BMC Genomics (2024)
-
RBBP6 maintains glioblastoma stem cells through CPSF3-dependent alternative polyadenylation
Cell Discovery (2024)
-
Cleavage and polyadenylation machinery as a novel targetable vulnerability for human cancer
Cancer Gene Therapy (2024)
-
RNA processing modification mediated subtypes illustrate the distinctive features of tumor microenvironment in hepatocellular carcinoma
Genes & Immunity (2024)
-
A distinct class of pan-cancer susceptibility genes revealed by an alternative polyadenylation transcriptome-wide association study
Nature Communications (2024)