Abstract
Climatic perturbation during the Pleistocene era has played a major role in plant evolutionary history by altering species distribution range. However, the relative roles of climatic and geographic factors in the distribution dynamics remain poorly understood; in particular, the edaphic endemics. In this paper, we examine the evolutionary history of two ultramafic primroses, Primula hidakana and Primula takedana. These species are ecologically and morphologically distinct with disjunct distributions on Hokkaido Island, Japan. Primula hidakana is found on various rocks in southern Hokkaido and P. takedana in serpentine areas in northern Hokkaido. We performed population genetics analyses on nuclear and chloroplast data sets and tested alternative phylogenetic models of divergence using approximate Bayesian computation (ABC) analyses. Nuclear microsatellite loci clearly distinguished the two sister taxa. In contrast, chloroplast sequence variations were shared between P. takedana and P. hidakana. ABC analyses based on nuclear data supported a secondary contact scenario involving asymmetrical gene flow from P. hidakana to P. takedana. Paleodistribution modeling also supported the divergence model, and predicted their latitudinal range shifts leading to past secondary contact. Our findings highlight the importance of the distribution dynamics during the Pleistocene climatic oscillations in the evolution of serpentine plants, and demonstrate that tight species cohesion between serpentine and nonserpentine sister taxa has been maintained despite past interspecific gene flow across soil boundaries.
Similar content being viewed by others
Introduction
Pleistocene climatic oscillations have played a key role in altering species distribution ranges and in providing opportunities for speciation through isolation (Comes and Kadereit 1998; Hewitt 2000, 2004). In other circumstances, repeated range shifts during climatic perturbation have also created opportunities for secondary contact between closely related species that were previously distributed in an allopatric manner. Accordingly, such distributional dynamics possibly result in interspecific hybridization (Rieseberg et al. 2003; Liu et al. 2014), reinforcement of reproductive barriers (Dobzhansky 1940; Kay and Schemske 2008), and transfer of adaptive alleles (Arnold et al. 2016). Interspecific gene exchange (i.e., introgression) in particular leads to homogenization during secondary contact of divergent gene pools, which may result in “reverse speciation” (Taylor et al. 2006). Therefore, gaining a greater insight into the latent roles of interspecific hybridization under Pleistocene climatic oscillations in a species’ evolutionary history is of general interest to both molecular ecologists and evolutionary biologists, as this can improve our understanding of how species are generated and maintained in nature.
Severe climatic perturbation may eliminate edaphic endemics (plants restricted to soils with unsuitable chemical or physical characteristics for many plants) from their original location, because their narrow edaphic niches limit successful migration (Van de Ven et al. 2007). On the other hand, because the niches of edaphic endemics are defined by soil type rather than climate, they may be able to endure climate change in their geographical ranges. Rather, several lines of evidence suggest that climatic cooling during glacial periods provides edaphic endemics with newly available but edaphically unsuitable habitats. Serpentine endemic plants, Arenaria norvegica and Minuartia laricifolia subsp. ophiolitica, both expanded out of serpentine outcrops when new nonserpentine habitats became available as the forest retreated during glacial periods (Rune 1953; Moore et al. 2013). These perspectives trigger another idea that Pleistocene climatic oscillations provided opportunities for secondary contact between edaphic endemics and their sister species, which were distributed allopatrically during the interglacial periods. Climate has long been thought to have played a key role in the evolutionary origin and persistence of serpentine plants and other edaphic endemics (Anacker and Harrison 2012; Anacker 2014; Rajakaruna 2018), yet the importance of past climatic oscillations in the evolution of edaphic endemics and their potential population dynamics, including interactions with their edaphically nonspecialized relatives, remains poorly understood.
Here, we examined the evolutionary history of recently diverged allopatric sister species, Primula hidakana (Miyabe and Kudo) Nakai and Primula takedana Tatew. (Primulaceae). We used phylogeographic and population genetics approaches to verify whether the Pleistocene climatic fluctuations drove interspecific gene flow between closely related taxa occurring in disjunct ultramafic habitats. Both species are diploid (2n = 24) self-incompatible perennial herbs that occur on the peridotite-rich metamorphic and serpentine belts of Hokkaido Island, Japan (Fig. 1a). Previous molecular phylogenies using both chloroplast and nuclear DNA showed that these two species have a robust sister relationship (Mast et al. 2001; Richards 2003; Yamamoto et al. 2017a), and inferred that P. takedana diverged recently (mid- to late Pleistocene) from P. hidakana (Yamamoto et al. 2017a). P. hidakana is found across the Hidaka ranges in southern Hokkaido (Fig. 1a), and occurs on wet, shaded rocky cliffs along a stream and a ridgeline in the mountains (Fig. 1b). Although the rock type is multifaceted, including peridotite, diabase, gneiss, and amphibolite (Watanabe 1971), P. hidakana does not occur in serpentine soils. Conversely, P. takedana is endemic to the denuded slopes of serpentine rocks partly weathered into clay along a stream in northern Hokkaido (Fig. 1a, c), and has a narrow and patchy distribution (Saito 1970). Their distribution ranges are separated by ∼200 km in a north–south direction (Fig. 1a), and no hybrids have been reported in their distribution ranges or in the interval zone. Although both species have been classified as ultramafic Primula (Kitamura 1950), there are differences between the species in relation to their morphological traits and physiological responses: nickel accumulation in P. takedana plants is much higher than in P. hidakana (Horie et al. 2000); and P. takedana has the typical morphological traits associated with adaptation to serpentine soils, including smaller flower size (Fig. 1d) and well-developed creeping rhizomes (Richards 2003); and the seed surface of P. takedana is covered with minute membrane organizations (Fig. 1e). These multifaceted observations indicate that P. takedana is the only species adapted to serpentine soils of the two ultramafic Primula, yet common garden studies testing the serpentine tolerance of P. hidakana are currently lacking.
a Location of Hokkaido Island and presumed distribution ranges of Primula takedana and P. hidakana. b Typical habitat of P. hidakana, Utabetsu-gawa stream, Erimo-cho, Horoizumi-gun, and Hokkaido. c Typical habitat of P. takedana, Nupuromapporo-gawa stream, Horonobe-cho, Teshio-gun, and Hokkaido. d Corolla morphologies of P. hidakana (pink) and P. takedana (white). e Seed morphologies of P. hidakana (left) and P. takedana (right)
In this study, we used nuclear microsatellite markers as well as multiple chloroplast DNA sequences to reconstruct the evolutionary history of the two ultramafic Primula in Hokkaido, Japan, utilizing an approximate Bayesian computational (ABC) approach (see Csilléry et al. 2012) and ecological niche modeling (ENM). The aims of this study were to elucidate the origin of serpentine endemic P. takedana, test alternative phylogenetic hypotheses for the divergence process, reconstruct historical range shifts of the currently disjunctive species, and better understand the role of past climatic changes in the speciation process of the edaphic endemic primroses.
Materials and methods
Population sampling, chloroplast DNA sequencing, and nuclear microsatellite genotyping
A total of 384 individuals from 24 populations (18 of P. hidakana and 6 of P. takedana) were collected throughout the entire range of the two species on Hokkaido Island, Japan (Fig. 2a). Fresh leaves from 16 individuals from each population were collected in silica gel, and genomic DNA was extracted from silica gel–dried leaf material following the cetyltrimethyl ammonium bromide method (Doyle 1990).
Polymorphisms in several plastid sequences using previous phylogeny (trnL-F, trnH-psbA, trnD-T, and rps16; Yamamoto et al. 2017a) were tested in four individuals of P. hidakana and P. takedana. This trial indicated that the two noncoding regions rps16 and trnD-T were the most informative markers. Each plastid region was amplified in four individuals per population using the primer pairs rpsF (5′-GTGGTAGAAAGCAACGTGCGACTT-3′)/rpsR2 (5′-TCGGGATCGAACATCAATTGCAAC-3′), and trnD (5′-AGGGCGGTACTCTAACCAA-3′)/trnT (5′-CGATGACTTACGCCTTACCA-3′) (Oxelman et al. 1997; Honjo et al. 2004). All sequencing experiments followed the methods described by Yamamoto et al. (2017a), and all new sequences have been deposited in the GenBank/DDBJ/EMBL database (LC330946–LC330969).
The nuclear genetic diversity of all 384 individuals was assessed using the following 12 microsatellite loci: Pok25, Pok32, Pok39, Pre02, Pre28, Pre51, Pre52, Pto06, Pto12, Pto21, Pto29, and Pto38 (Yamamoto et al. 2018a, 2018b). For all primer pairs, one of three different M13 universal sequences (5′-CACGACGTTGTAAAACGAC-3′, 5′-TGTGGAATTGTGA GCGG-3′, or 5′-CTATAGGGCACGCGTGGT-3′) and a pigtail sequence (5′-GTTTCTT-3′) were added to the 5′ end of each forward primer and the 3′ end of each reverse primer, respectively. The PCR reaction mix contained 5 µL Multiplex PCR Master Mix (Qiagen, Hilden, Germany), sterile water, 0.01 µM forward primer, 0.2 µM reverse primer, 0.1 µM fluorescence-labeled M13 primer, and 10–50 ng of template DNA in a total volume of 10 μL. The PCR amplification sequence was an initial 15 min of denaturation at 94 °C and 35 cycles consisting of 30 s of denaturation at 94 °C, 3 min of annealing at 60 °C, and 1 min of extension at 68 °C, with a final 20 min extension at 68 °C. Amplified products were mixed with highly deionized (Hi-Di) formamide and GeneScan 500 ROX size standard (Applied Biosystems, Foster City, CA, USA) and electrophoresed using an ABI 3130 Genetic Analyzer (Applied Biosystems). The fragment sizes were analyzed using GeneMapper (Applied Biosystems) and calibrated with AUTOBIN v0.9 (Salin 2013).
Data analyses of chloroplast haplotypes
Chloroplast regions rps16 and trnD-T were assembled and aligned with the Muscle program implemented in MEGA7 software (Kumar et al. 2016) using default settings with minor manual adjustment. The two plastid sequences were combined into a single sequence using SequenceMatrix v1.7.8 (Vaidya et al. 2011). Each individual was assigned to the different haplotypes using DnaSP v5 (Librado and Rozas 2009). DnaSP was also used to estimate molecular diversity indexes, including the number of haplotypes, haplotype diversity (h), and nucleotide diversity (π). Visualization of haplotype diversity distribution was performed via a median-joining haplotype network reconstructed using PopART v1.7 (Leigh and Bryant 2015). An analysis of molecular variance (AMOVA) was performed to partition variation within and among species and populations using Arlequin v3.5.1 (Excoffier and Lischer 2010). The significance of Φ-statistics was determined with 1000 permutations.
Data analyses, nuclear microsatellites
For all 12 nuclear microsatellite loci, the absence of linkage disequilibrium (LD), deviation from Hardy–Weinberg equilibrium (HWE), and the null allele frequency between loci in each population were detected using the program GENEPOP v4.2 (Raymond and Rousset 1995). LD and HWE tests were verified using a Markov chain method with 1000 dememorization steps, 100 batches, and 1000 iterations per batch. Null allele frequencies were estimated by the maximum-likelihood estimator based on the expectation–maximization algorithm (Dempster et al. 1977) with default settings. In addition, the effect of null alleles on population genetic differentiation (FST) was evaluated by comparing Weir’s FST (Weir 1996) before and after correction for null alleles using FreeNA software (Chapuis and Estoup 2007).
At the species level, for each locus and across loci, the following genetic diversity measurements were calculated using FSTAT v2.9.3 (Goudet 1995): A, number of alleles; AR, allelic richness; HO, observed heterozygosity within populations; HS, mean expected heterozygosity within populations; HT, total expected heterozygosity across populations; and FIS, fixation index. We also calculated the species-specific alleles and lineage-specific alleles that correspond to Bayesian clustering analysis (K = 2) in P. hidakana (see Fig. 5c).
At the population level, three genetic diversity statistics were calculated with FSTAT: AR, allelic richness; HO, observed heterozygosity; and HE, gene diversity (Nei 1987). In addition, the number of locally common alleles (i.e., frequency of more than 5% occurring in less than 25% of populations; AC) was calculated using GenAlEx v6.5 (Peakall and Smouse 2006). To graphically represent spatial patterns in population genetic diversity, smoothed interpolated maps were produced using an inverse distance weighting (IDW) interpolation method in Quantum GIS v2.18 (OSGeo, Beaverton, OR, USA) for the genetic diversity measurements of allelic richness, gene diversity, and locally common alleles. IDW in Quantum GIS was set at 2.0.
To evaluate genetic differentiation within species, we first calculated global FST (Weir and Cockrham 1984) and RST (Rousset 1996) using the SPAGeDi v1.5 (Hardy and Vekemans 2002). Index FST is based on only allele frequency variation, whereas RST is an index of genetic differentiation accounting for allele frequency and variance in allele size for microsatellite loci under a stepwise mutation model (Slatkin 1995). Following Hardy et al. (2003), SPAGeDi can be used to determine the relative importance of genetic drift versus mutation in genetic differentiation between populations by testing whether the observed RST value was significantly greater than the permuted RST values (pRST; 10,000 permutations). If the observed RST is significantly greater than pRST, a stepwise mutation model is the most likely explanation for genetic differentiation, suggesting a phylogeographic signal. Moreover, insignificant differences between RST and pRST suggest that allele size is not important, and that FST should be used instead of RST. Because RST was not significantly greater than pRST in this study (see Results), AMOVA was carried out based on FST using Arlequin.
Population genetic structures were examined using a Bayesian clustering analysis in STRUCTURE v2.3.2 (Pritchard et al. 2000). STRUCTURE analysis was conducted for all samples across the two species, and then the analysis was run separately within each species. Under an admixture model with correlated allele frequency, 20 independent simulations were run for each K (K = 1–20 for all samples, K = 1–15 for P. hidakana, and K = 1–10 for P. takedana) with 5 × 105 Markov chain Monte Carlo (MCMC) steps and a burn-in period of 105 iterations. For each dataset, the most likely value of K was determined by the ΔK method (Evanno et al. 2005) using STRUCTURE HARVESTER v0.6.94 (Earl and vonHoldt 2012). CLUMPAK (Kopelman et al. 2015) was used to average the outputs from multiple STRUCTURE runs and produce the graphical results.
To further detect the genetic grouping of microsatellite data, principal component analyses (PCA) were performed on a matrix of covariance values calculated from population allele frequency using GENODIVE (Meirmans and Van Tienderen 2004). We also used the Neighbor-Net algorithm (Bryant and Moulton 2004) to construct phylogenetic relationships between individuals based on microsatellite data. Construction of a Neighbor-Net phenogram based on the Bruvo genetic distance (Bruvo et al. 2004) was performed with SPLITSTREE4 v4.14.4 (Huson and Bryant 2005). In addition, spatial principal component analysis (sPCA) was performed to investigate genetic clines across the species distribution range using the R package adegenet v2.0.0 (Jombart 2008; Jombart and Ahmed 2011). sPCA was conducted in between- and within-species data sets using a distance-based connection network that assumes that all samples have at least one connection.
Evaluating speciation models by approximate Bayesian computation
We compared the following four hypothesized models of the speciation of P. hidakana and P. takedana (represented in Fig. 3) through analyses of the nuclear microsatellite data set using the ABC approach, as implemented in the ABCTOOLBOX software package (Wegmann et al. 2010): Model A, which assumed no gene flow throughout the divergence process (isolation model); Model B, which assumed constant gene flow since divergence (isolation-with-migration model); Model C, which assumed gene flow only in the initial phase of the divergence process (sympatric speciation model); and Model D, which assumed secondary contact after speciation and gene flow subsequent to an initial allopatric phase of divergence (isolation with secondary contact model). The parameters for prior population size (the size of each population remained constant), timing, and migration were assumed to have a log-uniform distribution (prior intervals for all model parameters are detailed in Supplementary Table 1).
Schematic representation of the four evolutionary models of speciation compared using approximate Bayesian computation (ABC): Model A, divergence without gene flow; Model B, isolation with migration; Model C, sympatric speciation; Model D, isolation with secondary contact model. Estimated posterior probabilities (PP) for each model are shown below each model (results based on δ = 0.001). Abbreviations are as follows: NT and NH, current effective population size of P. takedana and P. hidakana, respectively; NA, effective population size of their ancestor; T1, divergence time; T2, the time at which gene flow began; mH-T and mT-H, asymmetrical migration rate from P. hidakana to P. takedana and from P. takedana to P. hidakana, respectively
We examined nine statistics summarizing population genetic parameter information in the ABC analyses. These statistics were calculated using ARLSUMSTAT v3.5.2 (Excoffier and Lischer 2010) and are detailed in Supplementary Table 2. In the coalescent simulation step, one million simulations were generated for each model using the standard algorithm of ABCTOOLBOX and the program FASTSIMCOL2 v2.5.2.21 (Excoffier and Foll 2011). When simulating, the average mutation rate across loci was estimated to be 1.0 × 10−6–1.0 × 10−3 substitutions/site/year (Thuillet et al. 2002; Vigouroux et al. 2002; Resende-Moreira et al. 2017).
To evaluate which scenario best fit the observed summary statistics, the posterior probabilities (the marginal density of the model of interest divided by the sum of the marginal densities of all models) were estimated using the rejection method, and analyses were conducted with a range of tolerance values (δ = 0.01–0.001). For the best-fitting model, the sufficiency of the summary statistics and the robustness of our results were evaluated by posterior predictive checks (Csilléry et al. 2012). Population parameters in the best model were inferred with the neural network procedure (Blum and François 2010). Although accurate generation times (between seed germination and first flowering) of P. hidakana and P. takedana have not been recorded, the generation time of P. reinii var. rhodotricha, which is placed in the same section, has been estimated to be 2–3 years (Yamamoto et al. 2017b). Taking this into account, we assumed a generation time of three years for P. hidakana and P. takedana to estimate divergence time between the two species. All analyses described above were performed using the package abc (Csilléry et al. 2012) in R v3.4.1 (R Development Core Team 2017).
Bayesian population demographic analyses
In addition to evaluating speciation models using ABC analyses, we used the VarEff method implemented in the R package vareff (Nikolic and Chevalet 2014) to examine the past historically effective population sizes of P. takedana and P. hidakana. This method estimates prior changes in effective population size from microsatellite data (motif distance frequencies between alleles) by resolving coalescence theory and using an approximate likelihoods MCMC approach (Nikolic and Chevalet 2014). We used the prior parameter settings for each species (detailed in Supplementary Table 3) based on the results of our ABC analyses and a variety of preliminary runs. We ran each analysis under a two-phased mutation model with a proportion of 0.20 for multistep mutations (Peery et al. 2012), for 104 MCMC steps (NumberBatch = 10,000, LengthBatch = 10), sampling every 10 steps (SpaceBatch = 10), with an acceptance ratio of 0.25 (AccRate = 0.25). To assess time since changes in years, we used the estimated mutation rate 6.61 × 10−5 as estimated by our ABC results.
Ecological niche modeling
For ENM, we combined climate and nonclimate data coupled with species’ locality data at a spatial resolution of 30″ (∼1 km2). Coordinates from 33 localities for P. hidakana and 24 for P. takedana were obtained from our field collections and recent distribution research on P. takedana (Ito et al. 2016). The 19 bioclimatic variables for the current era (ca. 1950–2000) and the last interglacial (LIG; ca. 140–120 ka) were downloaded from the WorldClim database (http://www.worldclim.org/), whereas those for the last glacial maximum (LGM; ca. 21–18 ka) were obtained from two models; the Community Climate System Model (CCSM3: Collins et al. 2006) and the Model for Interdisciplinary Research on Climate (MIROC 3.2: Hasumi and Emori 2004). To avoid issues with multicollinearity (Dormann et al. 2013), six climatic variables (annual mean temperature, mean diurnal range as the mean of the monthly difference between the maximum temperature and the minimum temperature, maximum temperature of the warmest month, mean temperature of the warmest quarter, precipitation of the wettest quarter, and precipitation of the coldest quarter) were selected as uncorrelated climatic data. For nonclimate datasets, geology data were downloaded from a seamless digital geological map of Japan (1:200,000; Geological Survey of Japan 2015), whereas slope data were extracted from the National Land Numerical Information, Japan (http://nlftp.mlit.go.jp/).
ENMs were conducted using a maximum-entropy model implemented in MAXENT v3.3 (Phillips and Dudík 2008), which models presence-only species distribution data. All analyses were performed using the logistic output and a convergence threshold of 10−5 with 500 iterations, and then projected for the paleoclimate layers using both clamping and extrapolation methods (Merow et al. 2013). A total of 75% of the species’ records were included for training, and 25% for testing the model. Jackknife testing was conducted to assess the importance of each variable in the selected model. Model performances were evaluated using area under the curve (AUC) and true skill statistics (TSS; Allouche et al. 2006) based on 20 replicate runs. The 10th percentile training presence logistic threshold from MAXENT was used as the threshold for TSS because it produces accurate results when true absence data is lacking (Escalante et al. 2013).
We first ran the model for each species in MAXENT using all selected variables (i.e., “full model”) and then assessed the transferability of six climatic variables in the paleoclimate layers, because projecting species niche models into different time periods with possible nonanalog climate is not only liable to error, but also leads to unconvincing outcomes (e.g., Heikkinen et al. 2006; Fitzpatrick and Hargrove 2009). Based on the preliminary analyses, we removed the climatic variables that showed a broad extrapolated area and little contribution to the full model. Finally, we selected a model with two climatic variables (mean temperature of warmest quarter and precipitation of wettest quarter) and two nonclimate variables (geology and slope). The AUC and TSS values of the selected model were very similar to those of the full models. The selected model contained only very few or no values outside the training data range, which shows that no areas were strongly affected by variables beyond their training range.
The potential distribution maps of each species in three different time periods (current, LGM, and LIG) were illustrated with Quantum GIS. In addition, the overlap between the ranges of the two species at different time periods was computed using ENMTOOLS v1.3 (Warren et al. 2010) with probabilities greater than 0.307 (the 10th percentile training presence logistic threshold).
Results
Chloroplast haplotypes
The concatenated cpDNA sequences (rps16 and trnD-T) were aligned with a total length of 1355 bp, including 14 polymorphic sites and 19 insertion/deletion polymorphisms (indels). The chloroplast haplotype diversity (h) and nucleotide diversity (π) for each species are summarized in Table 1. The widespread P. hidakana had much higher levels of genetic diversity (h = 0.813; π = 3.59 × 10−3) than the geographically more restricted P. takedana (h = 0.359; π = 2.80 × 10−4).
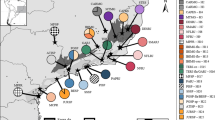
A total of 12 chloroplast haplotypes was identified among the 96 accessions including P. hidakana and P. takedana. Of these, nine haplotypes were specific to P. hidakana (H4–H12) and two to P. takedana (H2 and H3), whereas H1 was the most widespread and common haplotype for both species (Fig. 2). The haplotype network showed that two haplotypes specific to P. takedana (H2 and H3) and three haplotypes found in the northern part of the geographical distribution of P. hidakana (H4–H6) were linked to the main haplotype H1 by single steps in a “star-like” network (Fig. 2b). Conversely, haplotypes H8–H12 found in P. hidakana were six steps away from the main haplotype, H1, and were all confined to the southern part of their distribution range (Fig. 2b). The hierarchal AMOVAs revealed that only 7.0% of the genetic variation was partitioned between species with low and statistically nonsignificant genetic differentiation (ΦCT = 0.070; P > 0.05) (Table 2). On the other hand, high and significant among-population differentiation was observed for both species (ΦST = 0.870 for P. hidakana and ΦST = 0.792 for P. takedana).
Nuclear microsatellites
Each locus used in this study has sufficient polymorphism to conduct population genetic analyses (Supplementary Table 4). Furthermore, for all nuclear microsatellite markers, no evidence was found for a systematic departure from HWE at any particular locus in all populations, and no pairs of loci consistently showed LD in all populations. The estimated frequency of null alleles across populations varied from 0.02 to 0.31, with an average frequency of 0.06 in all 12 loci (Supplementary Table 5). However, no clear differences between uncorrected FST values and FST values corrected for all null alleles were found (Supplementary Table 6).
Genetic diversity measurements are summarized for all loci at the species (Table 1) and population levels (Supplementary Table 7). At the species level, all genetic diversity measurements (AR, HO, HS, and HT) were higher in P. hidakana (e.g., HS = 0.525) than in P. takedana (HS = 0.412). Furthermore, nonuniform spatial distributions of genetic diversity were observed for P. hidakana, in contrast to P. takedana, which consistently showed low genetic diversity among populations. The geographically northern (h2–h5) and southern (h13–h16) populations indicated higher genetic diversity than the central area of the distribution range of P. hidakana (Fig. 4a). The h3 population harbored the highest genetic diversity (AR = 4.92, HS = 0.603, and AC = 1.50), closely followed by the h14 population (AR = 5.17, HS = 0.591, and AC = 1.17) (Supplementary Table 7).
Geographical distribution of the genetic diversity among P. takedana (t1–t6) and P. hidakana (h1–h18) populations (a). The axis scores of the spatial principal component analysis (sPCA) (b); the locations of the samples are represented by circles that are colored according to a black to red gradient corresponding to the eigenvalues of the first (left) and second (right) axis of the sPCA, respectively
The number of species-specific alleles was larger in P. hidakana than that in P. takedana (Table 1), while in the three subdivisions investigated [P. takedana populations, and northern (h1–2, h4–7, and h11) and southern (h3, h8-18) P. hidakana populations], the southern P. hidakana population exhibited a greater number of private alleles per locus (= 3.33) compared with P. takedana (= 1.25) and northern P. hidakana populations (= 1.50).
Based on the FST–RST comparison, no significant difference was found in P. hidakana populations, whereas the RST value was significantly lower than the FST value in P. takedana populations (Table 1), suggesting that genetic drift is more important in this case. The AMOVAs using FST showed that 20.5% of the total nuclear microsatellite genetic variation was partitioned between species with moderate and significant genetic differentiation (ΦCT = 0.205, P < 0.05). In contrast to the pronounced levels of among-population genetic differentiation detected in P. hidakana (FST = 0.213, ΦST = 0.217), weak to moderate population differentiation was observed for P. takedana (FST = 0.079, ΦST = 0.083) (see Tables 1 and 2).
In the STRUCTURE analysis using all samples, the ΔK statistic peaked at K = 2, which indicates optimal division of the nuclear microsatellite data into two species (Supplementary Fig. 1a). However, some admixed individuals were also found in the P. hidakana populations (particularly in h3–5 and h14). These results were consistent throughout with other structuring results (see Fig. 5a). Further analysis indicated divergence of the northern populations of P. hidakana from the other populations at K = 3. For K = 4, some of the individuals in the cluster (blue or purple) in K = 3 were reassigned to a new cluster (green), which was distributed and scattered across P. hidakana populations. Further analyses within each species are shown in Fig. 5b, c. The ΔK statistics peaked at K = 3 and K = 2 for P. takedana and P. hidakana, respectively (Supplementary Fig. 1). Within P. takedana, although a weak genetic structure was detected, genetic admixture within three clusters was observed in all populations (Fig. 5b). Within P. hidakana, the two assigned clusters largely corresponded with the results of K = 3 using all samples across species (Fig. 5c).
The results of PCA and Neighbor-Net tree also supported the existence of two distinct genetic units corresponding to each species (Supplementary Fig. 2). sPCA distinguished the two species in the score of the first principle component, whereas the second axis differentiated the northern populations of P. hidakana from the others (Fig. 4b). At the species levels, although a latitudinal genetic cline was clearly detected within P. hidakana (Supplementary Fig. 6), several populations (e.g., h3 and h9) were discriminated from other northern populations. Conversely, such a spatial structure was weaker in the case of P. takedana.
Evaluating speciation models using approximate Bayesian computation
Based on ABC analyses, the isolation with secondary contact model (model D) was the most probable of the four proposed models for the speciation process of P. hidakana and P. takedana; the other three models received weaker support at all tolerance levels (Fig. 3, see also Supplementary Fig. 3). Posterior predictive checks confirmed that our observed summary statistics fell within distributions produced by parameter estimates from 10,000 randomly chosen accepted simulations in model D (Supplementary Fig. 4). Marginal posterior distributions of model parameters in model D are shown in Supplementary Fig. 5 and summarized in Table 3. Posterior parameter estimates suggested that P. hidakana and P. takedana diverged around 112 ka (95% highest posterior density interval, HPDI: 462–80 ka) and migration started at 45 ka (HPDI: 306–30 ka), assuming an average generation time of 3 years. The effective number of migrants per generation was unidirectional and quite high from P. hidakana to P. takedana (MH-T = 4.04, HPDI: 7.54 × 10−3 to 2.10 × 101), while the converse was almost zero. Although the modern effective population size of each species was much lower than the ancestral one (NA = 1.39 × 106; HPDI: 3.12 × 105 to 1.74 × 107), P. hidakana had a larger effective population size (NH = 1.30 × 104; HPDI: 4.39 × 104 to 6.21 × 105) than P. takedana (NT = 4.44 × 103; HPDI: 1.11 × 103 to 9.09 × 104).
Bayesian population demographic analyses
The demographic pattern outlined by VarEff indicates that the effective population sizes of each primrose were equivalent directly after isolation (Ne = 73,088 for P. hidakana, Ne = 51,493 for P. takedana). However, we also found that the effective population size of P. takedana has declined drastically over the past 25,000 years, whereas the curve of P. hidakana represents a gradual decline beginning 60,000 years ago. These results suggest that the timing and trend of the population decline was not identical between the two primroses; a severe population reduction in P. takedana began at or after the LGM, while P. hidakana experienced a gradual population decline during the LG period.
Ecological niche modeling
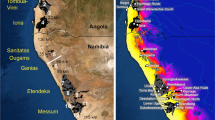
The selected models showed high predictive performance in both P. hidakana (AUC = 0.91 and TSS = 0.76) and P. takedana (AUC = 0.99 and TSS = 0.87), suggesting that the niche models for the two species capture their current actual distributions well (Fig. 6). The selected model’s internal jackknife test of variable importance showed that “geology” was the most important predictor for habitat distribution of P. takedana, whereas in addition to “geology”, “precipitation of wettest quarter” also contributed to the habitat prediction for P. hidakana (Supplementary Fig. 8).
Potential distribution ranges of P. takedana and P. hidakana at the present, last glacial maximum (LGM) and last interglacial (LIG) periods inferred by MAXENT. Darker colors represent areas with more suitable predicted conditions. CCSM, Community Climate System Model; MIROC, Model for Interdisciplinary Research on Climate
According to the models used for both species, their suitable habitats during the LIG period were consistently smaller than in the present climate, whereas the LGM projections differed depending on whether CCSM or MIROC climatic reconstructions were used (Fig. 6). The CCSM models predicted that suitable habitats for both species expanded between the LIG and LGM; the main distribution of P. takedana expanded and shifted towards the south, whereas some local expansions were also identified in the case of P. hidakana. In contrast, the MIROC models inferred that each species experienced contrasting distribution changes during the LGM period; the potential distribution range for P. hidakana largely contracted, whereas the one of P. takedana remained widespread throughout the serpentine belts of the Island. As in the present and LIG period, the computed range overlap between the two species during the LGM was zero in the MIROC model. However, their overlap based on the CCSM reconstructions was 0.21, which indicates that the ranges of the currently allopatric sister species overlapped slightly during the LGM period.
Discussion
Past interspecific gene flow between serpentine endemics and their progenitors throughout the Pleistocene climatic oscillations was not elucidated in previous studies; however, our population genetic analyses and ecological inferences were consistent in supporting the hypothesis that historical gene flow occurred between the currently disjunct progenitor-derivative species pair P. hidakana and P. takedana. ABC analyses supported a secondary contact model and inferred their population dynamics and recent evolutionary history. ENMs strongly supported the estimated speciation mode from our molecular data, and illustrated that their latitudinal range shifts in response to Pleistocene climatic changes resulted in secondary contact. Overall, our results imply that the late Pleistocene climatic oscillations contributed significantly to the alteration of the distribution ranges of serpentine endemics and created opportunities for secondary contact with their progenitors, leading to interspecific gene flow across soil boundaries.
Genetic diversity and structure
P. takedana was characterized by low genetic diversity and population differentiation in this study. Conversely, the nonuniform spatial distribution of genetic diversity and a phylogeographic signal were detected in P. hidakana. Genetic diversity was high in the northern and southern edges of their distribution range (Fig. 4a). These areas may mirror the persistence of P. hidakana during the Pleistocene climatic fluctuations and are candidate locations for their refugia. Thereby, the clear genetic structure along north–south direction (Fig. 5c) may be shaped by the refugial isolation during glacial cycles. This scenario can be reinforced by deep haplotype divergence between the northern and southern populations as observed in cpDNA (Fig. 2).
Meanwhile, Bayesian clustering using the nuclear dataset also detected heterogeneous populations within the northern area: although h3 and H8-10 populations were included in the same cpDNA haplotype group together with other northern populations (Fig. 2), these populations were clustered with geographically southern populations in nuclear microsatellites (Fig. 5c). Similar trends were also found in sPCA (Supplementary Fig. 6a). This complex phylogeographic pattern may be explained by the secondary contact between northern and southern lineages after colonization events. Considering the large number of private alleles found in the southern region, high allelic richness in the northern edge area (e.g., h3) might reflect recent gene flow from the southern to northern populations. Altogether, the current genetic structure of and spatial pattern of genetic diversity in P. hidakana was likely shaped by both historical isolation and recent gene flow between the northern and southern populations.
Origin of the serpentine endemic P. takedana
The serpentine-endemic P. takedana appears to have evolved recently from P. hidakana by vicariant separation. According to our results based on the chloroplast data set, there was extensive sharing of chloroplast haplotypes between species (Fig. 2) and no significant divergence (Table 2). This reflects the fact that P. takedana is likely to have originated recently. In fact, the ABC analyses suggested that the divergence occurred around 112 ka (Table 3), corresponding to the LIG period (Ono and Igarashi 1991). Although the time of divergence, as estimated by the ABC analyses, remain controversial due to the wide 95% HPDI (T1 = 463−80 ka; Table 3), the process of divergence had started before the major transition into the Würm glaciation at 75 ka (Ruddiman 1977).
Our ABC analysis supported a scenario with an initial phase of divergence without interspecific gene flow, implying that the initial splitting between species occurred in allopatry. In addition, despite the fact that P. takedana is rare and has lower genetic diversity than P. hidakana, our demographic reconstructions indicated that the two species have kept similar effective population size in the past (Supplementary Fig. 7). These results suggest that the initial phase of the divergence proceeded via a vicariant separation event rather than a peripheral isolate. Each primrose prefers cool and moist habitats (as discussed below), and it is likely that climatic amelioration during the mid to late Pleistocene period triggered speciation via vicariance range disjunction of their common ancestor that was once more widespread along the metamorphic belts of Hokkaido Island. The spatial distributions of cpDNA haplotype H1 and the star-like networks (see Fig. 2) plausibly reflect the range expansion of their ancestor, which occurred before the speciation events. Such vicariant separation scenarios have been hypothesized for the evolution of other serpentine plants (Kropf et al. 2008; Michl et al. 2010; Douglas et al. 2011; Moore et al. 2013). Our results, together with those of previous studies, highlight the relative role of Pleistocene climatic oscillations in the evolution of serpentine plants.
Secondary contact during the Pleistocene climatic oscillations
According to our ABC analyses, a model of isolation followed by secondary contact provides the best fit to our data. This result could be corroborated by chloroplast DNA analysis, which indicates that a major haplotype is shared across species (Fig. 2). Perhaps the indistinct species boundary implies the past introgression due to secondary contacts. On the contrary, a phylogeographic signature caused by secondary contact was unclear, particularly in the nuclear dataset. For example, genetic admixture at secondary contact zones often results in a genetic cline across species (Barton and Hewitt 1985). Although a few highly admixed individuals were found in several populations from the results of Bayesian clustering (e.g. h3 and h4), no clear genetic cline across species was detected in sPCA (Fig. 4b). In addition, secondary contacts between distinct lineages often lead to high local genetic diversity (e.g., Comps et al. 2001); however, past secondary contact between two Primula sister species could occur without leaving a trace on the spatial distribution of their genetic diversity, possibly due to a weak unidirectional gene flow. In our study, therefore, it would be premature to infer a speciation mode based on only the results of molecular data, which may lead to misinterpretation.
Nevertheless, our ecological inferences would support the secondary contact scenario. From the potential distributions estimated by the ENMs (Fig. 6), P. takedana obviously underwent a range expansion during the LGM, followed by a more recent contraction. Because the predicted distributions of both species were separated by a large region of unsuitable habitat when the climate was warm (i.e., at present and during the LIG period), this prediction makes it reasonable to postulate that the past secondary contact between the two species was caused by the expansion of their distributions and subsequent range overlap during a glacial period. Contrary to these inferences, the ENMs results for P. hidakana provide two contrasting range dynamics scenarios during the climatic cycles. The CCSM model indicated a wider LGM distribution of P. hidakana in comparison with the present and the LIG distributions, suggesting that the secondary contact was caused by the overlap during cool periods. In fact, our quantitative analysis predicts this range overlap between taxa during the LGM period. Conversely, according to the MIROC model, the distribution of P. hidakana was limited to their central geographical distribution area during the LGM period, and then spread from this area during the following postglacial warming period. This implies that the secondary contact occurred before or after the coolest part of the glacial period.
It is hard to distinguish the two contradictory scenarios suggested by the ENMs, because no palynological evidence has been found in either species. Nevertheless, our temporal framework provides further insights into their distributional dynamics. According to the time estimations provided by the ABC analyses, migration started prior to the LGM (T2 = 306—30 ka in 95% HPDI; Table 3), which contradicts the hypothesis that the secondary contact was caused by climatic amelioration after the coolest period, as suggested by the MIROC model. Moreover, the results of the Bayesian demographic inference also suggest interpretations of their distributional dynamics. If the species’ distribution range has changed over time, then evidence for such a change may be predicted based on the corresponding effective population sizes (Excoffier et al. 2009; Slatkin and Excoffier 2012). According to the results of our VarEff analyses (Supplementary Fig. 7), until at least the beginning of the LGM, the effective population sizes of P. takedana and P. hidakana were remarkably larger than at present. This implies that both species occupied wider distributional ranges during the LG period, as suggested by the CCSM model. Regardless of whether the CCSM or MIROC results are more accurate, however, our demographic and ecological inference are consistent with a migration during the cold period in the mid- to late Pleistocene, which provided opportunities for secondary contact between the two now disjunct species. In this case, the mode value of the time estimation suggests that secondary contact between species started during the LG period and the currently disjunctive and scattered distributions were shaped by the climatic amelioration between the LGM and the Holocene.
The serpentine endemic P. takedana is a clonal pioneer species (Saito 1970) and probably has high dispersal ability, as discussed below. Therefore, climatically harsh environments with relatively competition-free habitats during the glacial period may have facilitated the colonization of P. takedana into neighboring serpentine patches via edaphically unsuitable habitats. Considering the topographic characteristics of the Island, it is likely that, in addition to the Pleistocene climatic cycles, the latitudinal distribution of the serpentine belts and the absence of mountain chains dividing the lowlands would also have played an important role in the historical range shifts of P. takedana. Thus, our study suggests the relative roles of geological and climatic factors in the evolutionary origin and persistence of serpentine plants on Hokkaido Island, which harbors a wide variety of serpentine plants with high endemism (Brooks 1987).
Asymmetric gene flow possibly due to ecological differences
The estimated asymmetrical gene flow from P. hidakana to P. takedana during the past secondary contact phase is another key feature of this system. Such asymmetrical interspecific gene flow across soil boundaries was also reported in other edaphic endemic pairs, and was explained by pollinator behavior, dispersal ability or transfer of adaptive alleles (e.g. Douglas et al. 2011; Kay et al. 2018; Arnold et al. 2016). Although a thorough investigation of the explanations for asymmetrical migration is beyond the scope of this study, we conceived several potential explanations. First, the differences in corolla morphology between the two primroses may have contributed to the observed unidirectional gene flow. P. takedana flowers with funnel-shaped corollas (Fig. 1d) tend to expose their stigmas to various small insects, such as syrphid flies and short-tongued bees (including relatively ineffective pollinators of P. hidakana). On the other hand, P. hidakana flowers with narrow tubes (Fig. 1d) are mostly inaccessible to these short-tongued insects (Yamamoto et al. 2018c), which suggests that P. hidakana cannot receive P. takedana pollen. Hence, our findings suggest the possibility of the occurrence of asymmetrical prezygotic reproductive isolation due to corolla shape differentiation between recently diverged sister taxa. Such a unidirectional hybridization has frequently been reported between the co-occurring Primula species (e.g., Ma et al. 2014; Xie et al. 2017), possibly due to pollinator fauna differentiation.
Second, the differences in dispersal abilities among species may play an important role in promoting unidirectional gene flow. Serpentine plants tend to exhibit high genetic differentiation and low gene flow among populations (e.g. Wolf et al. 2000; Mengoni et al. 2003; Salmerón-Sánchez et al. 2014). In contrast, P. takedana is characterized by low population differentiation (Tables 1 and 2) despite their rarity and habitat isolation. As the natural habitats of P. takedana are strongly linked with riparian environments (Saito 1970; Ito et al. 2016), occasional seed-mediated gene flow by water (i.e., hydrochory) may be one of the most plausible explanations for the low genetic differentiation among populations. Interestingly, unlike closely related species, P. takedana exhibits seed buoyancy and the seed surface is densely covered with minute membrane organizations that may contribute to increased surface tension of the seeds (Fig. 1e) (Yamamoto, personal observation). It is thought that the relatively high dispersal ability of P. takedana may promote the spread of P. hidakana alleles into the entire population range of P. takedana from the past contact zones. Similar explanations have been discussed for the sympatric serpentine endemic Monardella species (Kay et al. 2018); introgression was predicted from M. stebinsii to M. folletti, which is characterized by lower FST than M. stebinsii.
Finally, this unidirectional migration may be explained by an adaptive process. Increase in metalliferous tolerant genotypes are usually believed to be rare in populations growing on nonmetalliferous soils (Kruckeberg 1951; Wu et al. 1975). Hence, alleles adapted for living in serpentine soils would be strongly selected against in nonserpentine soils. Conversely, migrant alleles from P. hidakana may have facilitated the adaptation of P. takedana to harsh environments and climatic perturbations, as has been found in Arabidopsis species (Arnold et al. 2016). Although we cannot provide further evidence to test these hypotheses, future analyses of large-scale genomic data sets, such as by restriction site-associated DNA sequencing, may shed light on the introgression and selection of alleles of sister species during the past secondary contact phase.
Conclusions
The predicted historical interspecific gene flow across soil boundaries provides a more complex scenario than a simple allopatric speciation model, which has been discussed in several edaphic endemics, and highlights the relative importance of Pleistocene climatic oscillations for the evolution of serpentine plants and their distributional dynamics. Although interspecific gene exchange has the potential to eliminate species boundaries (Taylor et al. 2006), divergence in reproductive traits and habitat preferences between P. hidakana and P. takedana might be key factors for the maintenance and reinforcement of their species integrity, as has been discussed in other plant groups (Matsuda et al. 2017; Kay et al. 2018). However, latent effects against the speciation process and local adaptation of ultramafic Primula arising from interspecific hybridization across soil boundaries remain unclear. Thus, studies of reproductive isolation between species, reciprocal transplantation, tests for local adaptation, and selection against hybrids are needed to better understand the roles of recent hybridization and speciation in the two ultramafic plants.
Data archiving
Sequence data have been submitted to GenBank/DDBJ/EMBL: accession numbers LC330946–LC330969. Microsatellite genotyping data have been submitted to Dryad: https://doi.org/10.5061/dryad.7r1p25r.
References
Allouche O, Tsoar A, Kadmon R (2006) Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J Appl Ecol 43:1223–1232
Anacker BL (2014) The nature of serpentine endemism. Am J Bot 101:219–224
Anacker BL, Harrison SP (2012) Climate and the evolution of serpentine endemism in California. Evol Ecol 26:1011–1023
Arnold BJ, Lahner B, DaCosta JM, Weisman CM, Hollister JD, Salt DE, Yant L (2016) Borrowed alleles and convergence in serpentine adaptation. Proc Natl Acad Sci USA 113:8320–8325
Barton NH, Hewitt GM (1985) Analysis of hybrid zones. Annu Rev Ecol Syst 16:113–148
Blum MG, François O (2010) Non-linear regression models for approximate Bayesian computation. Stat Comput 20:63–73
Brooks RR (1987) Serpentine and its vegetation: a multidisciplinary approach. Dioscorides Press, Portland
Bruvo R, Michiels NK, D’souza TG, Schulenburg H (2004) A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol Ecol 13:2101–2106
Bryant D, Moulton V (2004) Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21:255–265
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Collins WD, Bitz CM, Blackmon ML, Bonan GB, Bretherton CS, Carton JA, Kiehl JT (2006) The community climate system model version 3 (CCSM3). J Clim 19:2122–2143
Comes HP, Kadereit JW (1998) The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3:432–438
Comps B, Gömöry D, Letouzey J, Thiébaut B, Petit RJ (2001) Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics 157:389–397
Csilléry K, François O, Blum MG (2012) ABC: an R package for approximate Bayesian computation (ABC). Methods Ecol Evol 3:475–479
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc 39:1–38
Dobzhansky T (1940) Speciation as a stage in evolutionary divergence. Am Nat 74:312–321
Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, Münkemüller T (2013) Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36:27–46
Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Douglas NA, Wall WA, XIANG QYJ, Hoffmann WA, Wentworth TR, Gray JB, Hohmann MG (2011) Recent vicariance and the origin of the rare, edaphically specialized Sandhills lily, Lilium pyrophilum (Liliaceae): evidence from phylogenetic and coalescent analyses. Mol Ecol 20:2901–2915
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Escalante T, Rodríguez-Tapia G, Linaje M, Illoldi-Rangel P, González-López R (2013) Identification of areas of endemism from species distribution models: threshold selection and Nearctic mammals. TIP Rev Espec Cienc Quím Biol 16:5–17
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Foll M, Petit RJ (2009) Genetic consequences of range expansions. Annu Rev Ecol Evol Syst 40:481–501
Excoffier L, Foll M (2011) Fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex scenarios. Bioinfomatics 27:1332–1334
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fitzpatrick MC, Hargrove WW (2009) The projection of species distribution models and the problem of non-analog climate. Biodivers Conserv 18:2255
Goudet J (1995) FSTAT: a computer program to calculate F-statistics. Heredity 86:485–486
Geological Survey of Japan (2015) Seamless digital geological map of Japan 1: 200,000. Geological Survey of Japan, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan
Hardy OJ, Charbonnel N, Fréville H, Heuertz M (2003) Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163:1467–1482
Hasumi H, Emori S (2004) K-1 coupled GCM (MIROC) description. Center for Climate System Research, University of Tokyo, Tokyo
Heikkinen RK, Luoto M, Araújo MB, Virkkala R, Thuiller W, Sykes MT (2006) Methods and uncertainties in bioclimatic envelope modelling under climate change. Prog Phys Geogr 30:751–777
Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B Biol Sci 359:183–195
Honjo M, Ueno S, Tsumura Y, Washitani I, Ohsawa R (2004) Phylogeographic study based on intraspecific sequence variation of chloroplast DNA for the conservation of genetic diversity in the Japanese endangered species Primula sieboldii. Biol Conserv 120:211–220
Horie K, Mizuno N, Nosaka S (2000) Characteristics of nickel accumulation in native plants growing in ultramafic rock areas in Hokkaido. Soil Sci Plant Nutr 46:853–863
Huson DH, Bryant D (2005) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Ito K, Takahashi H, Hayakashi S, Okuda A, Wada K, Kikuchi S (2016) Survey on distribution of Primula takedana and Japonolirion osense in Teshio Experimental Forest and Nakagawa Experimental Forest of Hokkaido University. Technical Report for Boreal Forest Conservation of the Hokkaido University Forests, 33, 1–3. (In Japanese)
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Ahmed I (2011) Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Kay KM, Schemske DW (2008) Natural selection reinforces speciation in a radiation of neotropical rainforest plants Evolution 62:2628–2642
Kay KM, Woolhouse S, Smith BA, Pope NS, Rajakaruna N (2018) Sympatric serpentine endemic Monardella (Lamiaceae) species maintain habitat differences despite hybridization. Mol Ecol 27:2302–2316
Kitamura S (1950) Adaptation and isolation on the serpentine areas. Acta Phytotaxon Geobot 12:178–185. (In Japanese)
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191
Kropf M, Comes HP, Kadereit JW (2008) Causes of the genetic architecture of south-west European high mountain disjuncts. Plant Ecol Divers 1:217–228
Kruckeberg AR (1951) Intraspecific variability in the response of certain native plant species to serpentine soil. Am J Bot 38:408–419
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Leigh JW, Bryant D (2015) Popart: full‐feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Librado P, Rozas J (2009) DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu B, Abbott RJ, Lu Z, Tian B, Liu J (2014) Diploid hybrid origin of Ostryopsis intermedia (Betulaceae) in the Qinghai‐Tibet Plateau triggered by Quaternary climate change. Mol Ecol 23:3013–3027
Ma Y, Xie W, Tian X, Sun W, Wu Z, Milne R (2014) Unidirectional hybridization and reproductive barriers between two heterostylous primrose species in north-west Yunnan, China. Ann Bot 113:763–775
Mast AR, Kelso S, Richards AJ, Lang DJ, Feller DM, Conti E (2001) Phylogenetic relationships in Primula L. and related genera (Primulaceae) based on noncoding chloroplast DNA. Int J Plant Sci 162:1381–1400
Matsuda J, Maeda Y, Nagasawa J, Setoguchi H (2017) Tight species cohesion among sympatric insular wild gingers (Asarum spp. Aristolochiaceae) on continental islands: highly differentiated floral characteristics versus undifferentiated genotypes. PLoS ONE 12:e0173489
Mengoni A, Gonnelli C, Brocchini E, Galardi F, Pucci S, Gabbrielli R, Bazzicalupo M (2003) Chloroplast genetic diversity and biogeography in the serpentine endemic Ni‐hyperaccumulator Alyssum bertolonii. New Phytol 157:349–356
Merow C, Smith MJ, Silander Jr JA (2013) A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography 36:1058–1069
Meirmans PG, Van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Michl T, Huck S, Schmitt T, Liebrich A, Haase P, Büdel B (2010) The molecular population structure of the tall forb Cicerbita alpina (Asteraceae) supports the idea of cryptic glacial refugia in central Europe. Bot J Linn Soc 164:142–154
Moore AJ, Merges D, Kadereit JW (2013) The origin of the serpentine endemic Minuartia laricifolia subsp. ophiolitica by vicariance and competitive exclusion. Mol Ecol 22:2218–2231
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, NY
Nikolic N, Chevalet C (2014) Detecting past changes of effective population size. Evol Appl 7:663–681
Hardy OJ, Vekemans X (2002) spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Ono Y, Igarashi Y (1991) Natural history of Hokkaido. Hokkaido University Press, Hokkaido, Japan
Oxelman B, Lidén M, Berglund D (1997) Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Syst Evol 206:393–410
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peery MZ, Kirby R, Reid BN, Stoelting R, PALSBØLL PJ (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol 21:3403–3418
Phillips SJ, Dudík M (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31:161–175
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team (2017) R: a Language and Environment for Statistical Computing. https://www.R-project.org/.
Rajakaruna N (2018) Lessons on evolution from the study of edaphic specialization. Bot Rev 84:39–78
Raymond M, Rousset F (1995) GENEPOP (Version1.2): population genetics software for exact tests and ecumenicism. Heredity 86:248–249
Richards AJ (2003) Primula. B. T. Batsford Press, London
Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Lexer C (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216
Resende-Moreira LC, Ramos ACS, Scliar MO, Silva RM, Azevedo VC, Ciampi AY, Lovato MB (2017) Gene flow between vicariant tree species: insights into savanna-forest evolutionary relationships. Tree Genet Genomes 13:36
Rousset F (1996) Equilibrium values of measures of population subdivision for stepwise mutation processes. Genetics 142:1357–1362
Ruddiman WF (1977) Late Quaternary deposition of ice-rafted sand in the subpolar North Atlantic (lat 40° to 65° N). Geol Soc Am Bull 88:1813–1827
Rune O (1953) Notes on the flora of the Gaspé Peninsula. Sven Bot Tidskr 48:117–135
Salmerón-Sánchez E, Merlo ME, Medina-Cazorla JM, Pérez-García FJ, Martínez-Hernández F, Garrido-Becerra JA, Mota JF (2014) Variability, genetic structure and phylogeography of the dolomitophilous species Convolvulus boissieri (Convolvulaceae) in the Baetic ranges, inferred from AFLPs, plastid DNA and ITS sequences. Bot J Linn Soc 176:506–523
Saito S (1970) A study on the environment of teshio primrose (Primula takedana Tatewaki). Res Bull Coll Exp For Hokkaido Univ 27:49–62
Salin F (2013) AutoBin. https://www6.bordeaux-aquitaine.inra.fr/biogeco/Production-scientifique/Logiciels/Autobin. Accessed Sept 2016
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Slatkin M, Excoffier L (2012) Serial founder effects during range expansion: a spatial analog of genetic drift. Genetics 191:171–181
Taylor EB, Boughman JW, Groenenboom M, Sniatynski M, Schluter D, Gow JL (2006) Speciation in reverse: morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol Ecol 15:343–355
Thuillet AC, Bru D, David J, Roumet P, Santoni S, Sourdille P, Bataillon T (2002) Direct estimation of mutation rate for 10 microsatellite loci in durum wheat, Triticum turgidum (L.) Thell. ssp durum desf. Mol Biol Evol 19:122–125
Vaidya G, Lohman DJ, Meier R (2011) SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics 27:171–180
Van de Ven CM, Weiss SB, Ernst WG (2007) Plant species distributions under present conditions and forecasted for warmer climates in an arid mountain range. Earth Interact 11:1–33
Vigouroux Y, Jaqueth JS, Matsuoka Y, Smith OS, Beavis WD, Smith JSC, Doebley J (2002) Rate and pattern of mutation at microsatellite loci in maize. Mol Biol Evol 19:1251–1260
Watanabe S (1971) Phytogeographical studies of the alpine plants (vascular plants) on the Hidaka-Yubari ranges, Hokkaido. Mem Natl Mus Nat Sci 4:95
Warren DL, Glor RE, Turelli M (2010) ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33:607–611
Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L (2010) ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinform 11:116
Weir BS (1996) Genetic data analysis II: methods for discrete population genetic data. Sinauer Associates, Sounderland, MA
Weir BS, Cockerham CC (1984) Estimating F‐statistics for the analysis of population structure. Evolution 38:1358–1370
Wolf AT, Howe RW, Hamrick JL (2000) Genetic diversity and population structure of the serpentine endemic Calystegia collina (Convolvulaceae) in Northern California. Am J Bot 87:1138–1146
Wu L, Bradshaw AD, Thurman DA (1975) The potential for the evolution of heavy‐metal tolerance in plants. III. The rapid evolution of copper tolerance in Agrostis stolonifera. Heredity 34:165–187
Xie Y, Zhu X, Ma Y, Zhao J, Li L, Li Q (2017) Natural hybridization and reproductive isolation between two Primula species. J Integr Plant Biol 59:526–530
Yamamoto M, Ohtani M, Kurata K, Setoguchi H (2017a) Contrasting evolutionary processes during Quaternary climatic changes and historical orogenies: a case study of the Japanese endemic primroses Primula sect reinii. Ann Bot 120:943–954
Yamamoto M, Setoguchi H, Kurata K (2017b) Conservation genetics of an ex situ population of Primula reinii var. rhodotricha, an endangered primrose endemic to Japan on a limestone mountain. Conserv Genet 18:1141–1150
Yamamoto M, Handa Y, Aihara H, Setoguchi H (2018a) Development and characterization of 24 microsatellite markers in Primula tosaensis, an endangered primrose, using MiSeq. Plant Species Biol 33:77–80
Yamamoto M, Handa Y, Aihara H, Setoguchi H (2018b) Development and characterization of 43 microsatellite markers for the critically endangered primrose Primula reinii using MiSeq sequencing. Plant Divers 40:41–44
Yamamoto M, Horita K, Takahashi D, Murai Y, Setoguchi H (2018c) Floral morphology and pollinator fauna of sister species Primula takedana and P. hidakana in Hokkaido island, Japan. Bull Natl Sci Mus Ser B 44:97–103
Acknowledgements
This work was financially supported by Grants-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science (#24247013, #26304013 and #19K16219) and the Field Science Centre for Northern Biosphere, Hokkaido University (#201604D). We are grateful to S. Sakaguchi, Y. Yamasaki, K. Ito, S. Umezawa and K. Ueno for assistance with sample collection and their valuable comments on this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yamamoto, M., Takahashi, D., Horita, K. et al. Speciation and subsequent secondary contact in two edaphic endemic primroses driven by Pleistocene climatic oscillation. Heredity 124, 93–107 (2020). https://doi.org/10.1038/s41437-019-0245-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-019-0245-8