Abstract
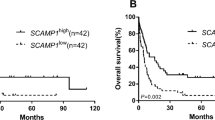
Genetic abnormalities are more frequently viewed as prognostic markers in acute myeloid leukemia (AML) in recent years. Fucosylation, catalyzed by fucosyltransferases (FUTs), is a post-translational modification that widely exists in cancer cells. However, the expression and clinical implication of the FUT family (FUT1-11) in AML has not been investigated. From the Cancer Genome Atlas database, a total of 155 AML patients with complete clinical characteristics and FUT1-11 expression data were included in our study. In patients who received chemotherapy alone showed that high expression levels of FUT3, FUT6, and FUT7 had adverse effects on event-free survival (EFS) and overall survival (OS) (all P < 0.05), whereas high FUT4 expression had favorable effects on EFS and OS (all P < 0.01). However, in the allogeneic hematopoietic stem cell transplantation (allo-HSCT) group, we only found a significant difference in EFS between the high and low FUT3 expression subgroups (P = 0.047), while other FUT members had no effect on survival. Multivariate analysis confirmed that high FUT4 expression was an independent favorable prognostic factor for both EFS (HR = 0.423, P = 0.001) and OS (HR = 0.398, P < 0.001), whereas high FUT6 expression was an independent risk factor for both EFS (HR = 1.871, P = 0.017) and OS (HR = 1.729, P = 0.028) in patients who received chemotherapy alone. Moreover, we found that patients with low FUT4 and high FUT6 expressions had the shortest EFS and OS (P < 0.05). Our study suggests that high expressions of FUT3/6/7 predict poor prognosis, high FUT4 expression indicates good prognosis in AML; FUT6 and FUT4 have the best prognosticating profile among them, but their effects could be neutralized by allo-HSCT.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–52.
Zhang J, Gu Y, Chen B. Mechanisms of drug resistance in acute myeloid leukemia. Onco Targets Ther. 2019;12:1937–45.
Mrozek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30:4515–23.
Zhu YM, Wang PP, Huang JY, Chen YS, Chen B, Dai YJ, et al. Gene mutational pattern and expression level in 560 acute myeloid leukemia patients and their clinical relevance. J Transl Med. 2017;15:178.
Wakita S, Yamaguchi H, Omori I, Terada K, Ueda T, Manabe E, et al. Mutations of the epigenetics-modifying gene (DNMT3a, TET2, IDH1/2) at diagnosis may induce FLT3-ITD at relapse in de novo acute myeloid leukemia. Leukemia. 2013;27:1044–52.
Su L, Gao SJ, Li W, Tan YH, Cui JW, Hu RP. NPM1, FLT3-ITD, CEBPA, and c-kit mutations in 312 Chinese patients with de novo acute myeloid leukemia. Hematology. 2014;19:324–8.
Fasan A, Haferlach C, Alpermann T, Jeromin S, Grossmann V, Eder C, et al. The role of different genetic subtypes of CEBPA mutated AML. Leukemia. 2014;28:794–803.
Shamaa S, Laimon N, Aladle DA, Azmy E, Elghannam DM, Salem DA, et al. Prognostic implications of NPM1 mutations and FLT3 internal tandem duplications in Egyptian patients with cytogenetically normal acute myeloid leukemia. Hematology. 2014;19:22–30.
Cheng Z, Dai Y, Pang Y, Jiao Y, Zhao H, Zhang Z, et al. Enhanced expressions of FHL2 and iASPP predict poor prognosis in acute myeloid leukemia. Cancer Gene Ther. 2019;26:17–25.
Zhang L, Li R, Hu K, Dai Y, Pang Y, Jiao Y, et al. Prognostic role of DOK family adapters in acute myeloid leukemia. Cancer Gene Ther. https://doi.org/10.1038/s41417-018-0052-z.
Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158r–184r.
Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Marth JD, et al. Symbol nomenclature for glycan representation. Proteomics. 2009;9:5398–9.
Cheng L, Luo S, Jin C, Ma H, Zhou H, Jia L. FUT family mediates the multidrug resistance of human hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell Death Dis. 2013;4:e923.
Liu YC, Yen HY, Chen CY, Chen CH, Cheng PF, Juan YH, et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc Natl Acad Sci USA. 2011;108:11332–7.
Liang L, Gao C, Li Y, Sun M, Xu J, Li H, et al. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017;8:e2968.
Weston BW, Hiller KM, Mayben JP, Manousos GA, Bendt KM, Liu R, et al. Expression of human alpha(1,3)fucosyltransferase antisense sequences inhibits selectin-mediated adhesion and liver metastasis of colon carcinoma cells. Cancer Res. 1999;59:2127–35.
Iwamori M, Tanaka K, Kubushiro K, Lin B, Kiguchi K, Ishiwata I, et al. Alterations in the glycolipid composition and cellular properties of ovarian carcinoma-derived RMG-1 cells on transfection of thealpha1,2-fucosyltransferase gene. Cancer Sci. 2005;96:26–30.
Zhao Y, Lin B, Hao YY. The effects of Lewis(y) antigen content on drug resistance tocarboplatin in ovarian cancer line RMG-I. Prog Biochem Biophys. 2008;35:1175–82.
Yang XS, Zhang ZB, Jia SA, Liu YJ, Wang XQ, Yan Q. Overexpression of fucosyltransferase IV in A431 cell line increases cell proliferation. Int J Biochem. Cell B. 2007;39:1722–30.
Wang QY, Guo P, Duan LL, Shen ZH, Chen HL. alpha-1,3-Fucosyltransferase-VII stimulates the growth of hepatocarcinoma cells via the cyclin-dependent kinase inhibitor p27Kip1. Cell Mol Life Sci. 2005;62:171–8.
Hiller KM, Mayben JP, Bendt KM, Manousos GA, Senger K, Cameron HS, et al. Transfection of alpha(1,3)fucosyltransferase antisense sequences impairs the proliferative and tumorigenic ability of human colon carcinoma cells. Mol Carcinog. 2000;27:280–8.
Kang X, Wang N, Pei C, Sun L, Sun R, Chen J, et al. Glycan-related gene expression signatures in human metastatic hepatocellular carcinoma cells. Exp Ther Med. 2012;3:415–22.
Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74.
Cheson BD, Bennett JM, Kopecky KJ, Buchner T, Willman CL, Estey EH, et al. Revised recommendations of the international working group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003;21:4642–9.
Desiderio V, Papagerakis P, Tirino V, Zheng L, Matossian M, Prince ME, et al. Increased fucosylation has a pivotal role in invasive and metastatic properties of head and neck cancer stem cells. Oncotarget. 2015;6:71–84.
Gao HF, Wang QY, Zhang K, Chen LY, Cheng CS, Chen H, et al. Overexpressed N-fucosylation on the cell surface driven by FUT3, 5, and 6 promotes cell motilities in metastatic pancreatic cancer cell lines. Biochem Biophys Res Commun. 2019;511:482–9.
Zhan L, Chen L, Chen Z. Knockdown of FUT3 disrupts the proliferation, migration, tumorigenesis and TGF-β induced EMT in pancreatic cancer cells. Oncol Lett. 2018;16:924–30.
Vasconcelos JLD, Ferreira SD, de Lima ALR, Rego MJBD, Bandeira ARG, Cavalcanti CDB, et al. Comparing the Immunoexpression of FUT3 and FUT6 between Prostatic Adenocarcinoma and Benign Prostatic Hyperplasia. Acta Histochem Cytoc. 2013;46:105–9.
Petretti T, Kemmner W, Schulze B, Schlag PM. Altered mRNA expression of glycosyltransferases in human colorectal carcinomas and liver metastases. Gut. 2000;46:359–66.
do Nascimento JC, Ferreira Sde A, Vasconcelos JL, da Silva-Filho JL, Barbosa BT, Bezerra MF, et al. Fut3 role in breast invasive ductal carcinoma: Investigating its gene promoter and protein expression. Exp Mol Pathol. 2015;99:409–15.
Li D, Sun H, Bai G, Wang W, Liu M, Bao Z, et al. alpha-1,3-Fucosyltransferase-VII siRNA inhibits the expression of SLex and hepatocarcinoma cell proliferation. Int J Mol Med. 2018;42:2700–8.
Liang JX, Gao W, Cai L. Fucosyltransferase VII promotes proliferation via the EGFR/AKT/mTOR pathway in A549 cells. Onco Targets Ther. 2017;10:3971–8.
Pan S, Liu Y, Liu Q, Xiao Y, Liu B, Ren X, et al. HOTAIR/miR-326/FUT6 axis facilitates colorectal cancer progression through regulating fucosylation of CD44 via PI3K/AKT/mTOR pathway. Biochim Biophys Acta, Mol Cell Res. 2019;1866:750–60.
Li J, Guillebon AD, Hsu JW, Barthel SR, Dimitroff CJ, Lee YF, et al. Human fucosyltransferase 6 enables prostate cancer metastasis to bone. Br J Cancer. 2013;109:3014–22.
Li N, Liu Y, Miao Y, Zhao L, Zhou H, Jia L. MicroRNA-106b targets FUT6 to promote cell migration, invasion, and proliferation in human breast cancer. IUBMB life. 2016;68:764–75.
Taniguchi A, Suga R, Matsumoto K. Expression and transcriptional regulation of the humanalpha1, 3-fucosyltransferase 4 (FUT4) gene in myeloid and colon adenocarcinoma cell lines. Biochem Biophys Res Commun. 2000;273:370–6.
Li Y, Sun Z, Liu B, Shan Y, Zhao L, Jia L. Tumor-suppressive miR-26a and miR-26b inhibit cell aggressiveness by regulating FUT4 in colorectal cancer. Cell Death Dis. 2017;8:e2892.
Ogawa J, Inoue H, Koide S. Expression of alpha-1,3-fucosyltransferase type IV and VII genes is related to poor prognosis in lung cancer. Cancer Res. 1996;56:325–9.
Schoen MW, Woelich SK, Braun JT, Reddy DV, Fesler MJ, Petruska PJ, et al. Acute myeloid leukemia induction with cladribine: Outcomes by age and leukemia risk. Leuk Res. 2018;68:72–78.
Bacher U, Haferlach C, Alpermann T, Kern W, Schnittger S, Haferlach T. Comparison of genetic and clinical aspects in patients with acute myeloid leukemia and myelodysplastic syndromes all with more than 50% of bone marrow erythropoietic cells. Haematologica. 2011;96:1284–92.
Kumar D, Mehta A, Panigrahi MK, Nath S, Saikia KK. DNMT3A (R882) mutation features and prognostic effect in acute myeloid leukemia in coexistent with NPM1 and FLT3 mutations. Hematol Oncol Stem Cell Ther. 2018;11:82–89.
Welch JS. Patterns of mutations in TP53 mutated AML. Best Pr Res Clin Haematol. 2018;31:379–83.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81500118 and 61501519), the China Postdoctoral Science Foundation funded project (2016M600443), and Jiangsu Province Postdoctoral Science Foundation funded project (1701184B).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dai, Y., Cheng, Z., Pang, Y. et al. Prognostic value of the FUT family in acute myeloid leukemia. Cancer Gene Ther 27, 70–80 (2020). https://doi.org/10.1038/s41417-019-0115-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-019-0115-9
This article is cited by
-
Alpha-(1,6)-fucosyltransferase (FUT8) affects the survival strategy of osteosarcoma by remodeling TNF/NF-κB2 signaling
Cell Death & Disease (2021)
-
FUT6 deficiency compromises basophil function by selectively abrogating their sialyl-Lewis x expression
Communications Biology (2021)