Abstract
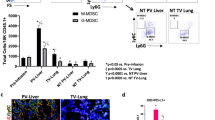
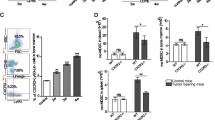
Myeloid derived suppressor cells (MDSCs) play key roles in cancer development. Accumulation of peripheral-blood MDSCs (PB-MDSCs) corresponds to the progression of various cancers, but provides only a crude indicator. We aimed toward identifying changes in the transcriptional profile of PB-MDSCs in response to tumor growth. CT26 colon cancer cells and B16 melanoma cells (106) were inoculated into peritoneal cavities of BALB/c mice and subcutaneously to C57-black mice, respectively. The circulating levels and global transcriptional patterns of PB CD11b+Ly6g+ MDSCs were assessed in control mice, and 4, 8, and 11 days following tumor cell inoculation. Although a significant accumulation of PB-MDSCs was demonstrated only 11 days following tumor induction, a pronounced transcriptional response was identified already on day 4 while the tumor was ~1 mm in size. Further transcriptional changes correlated with different stages of tumor growth. Key MDSC genes and canonical signaling pathways were activated along tumor progression. This phenomenon was demonstrated in both cancer models, and a consensus set of 817 genes, involved in myeloid cell recruitment and angiogenesis, was identified. The data suggest that the transcriptional signatures of PB-MDSC may serve as markers for tumor progression, as well as providing potential targets for future therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Folkman J, Kalluri R. Cancer without disease. Nature. 2004;427:787.
Gimbrone MA, Jr, Leapman SB, Cotran RS, Folkman J. Tumor dormancy in vivo by prevention of neovascularization. J Exp Med. 1972;136:261–76.
Folkman J, Klagsbrun M. Angiogenic factors. Science. 1987;235:442–7.
Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, et al. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell. 2004;6:409–21.
Sica A, Porta C, Amadori A, Pasto A. Tumor-associated myeloid cells as guiding forces of cancer cell stemness. Cancer Immunol Immunother. 2017;66:1025–36.
Yan HH, Pickup M, Pang Y, Gorska AE, Li Z, Chytil A, et al. Gr-1+CD11b+ myeloid cells tip the balance of immune protection to tumor promotion in the premetastatic lung. Cancer Res. 2010;70:6139–49.
Okazaki T, Ebihara S, Asada M, Kanda A, Sasaki H, Yamaya M. Granulocyte colony-stimulating factor promotes tumor angiogenesis via increasing circulating endothelial progenitor cells and Gr1+CD11b+ cells in cancer animal models. Int Immunol. 2006;18:1–9.
Shojaei F, Ferrara N. Refractoriness to antivascular endothelial growth factor treatment: role of myeloid cells. Cancer Res. 2008;68:5501–4.
Shojaei F, Wu X, Malik AK, Zhong C, Baldwin ME, Schanz S, et al. Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+Gr1+ myeloid cells. Nat Biotechnol. 2007;25:911–20.
Zhao L, Lim SY, Gordon-Weeks AN, Tapmeier TT, Im JH, Cao Y, et al. Recruitment of a myeloid cell subset (CD11b/Gr1 mid) via CCL2/CCR2 promotes the development of colorectal cancer liver metastasis. Hepatology. 2013;57:829–39.
Panni RZ, Sanford DE, Belt BA, Mitchem JB, Worley LA, Goetz BD, et al. Tumor-induced STAT3 activation in monocytic myeloid-derived suppressor cells enhances stemness and mesenchymal properties in human pancreatic cancer. Cancer Immunol Immunother. 2014;63:513–28.
Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res. 2011;9:133–48.
Diaz-Montero CM, Finke J, Montero AJ. Myeloid-derived suppressor cells in cancer: therapeutic, predictive, and prognostic implications. Semin Oncol. 2014;41:174–84.
Gabrilovich DI. Myeloid-derived suppressor cells. Cancer Immunol Res. 2017;5:3–8.
Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–20.
Gabitass RF, Annels NE, Stocken DD, Pandha HA, Middleton GW. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol Immunother. 2011;60:1419–30.
Sun HL, Zhou X, Xue YF, Wang K, Shen YF, Mao JJ, et al. Increased frequency and clinical significance of myeloid-derived suppressor cells in human colorectal carcinoma. World J Gastroenterol. 2012;18:3303–9.
Khaled YS, Ammori BJ, Elkord E. Increased levels of granulocytic myeloid-derived suppressor cells in peripheral blood and tumour tissue of pancreatic cancer patients. J Immunol Res. 2014;2014:879897.
Zhang B, Wang Z, Wu L, Zhang M, Li W, Ding J, et al. Circulating and tumor-infiltrating myeloid-derived suppressor cells in patients with colorectal carcinoma. PLoS ONE. 2013;8:e57114.
Choi J, Maeng HG, Lee SJ, Kim YJ, Kim DW, Lee HN, et al. Diagnostic value of peripheral blood immune profiling in colorectal cancer. Ann Surg Treat Res. 2018;94:312–21.
Jin G, Zhang Y, Chang X, Zhang Y, Xu J, Wei M, et al. Increased percentage of mo-MDSCs in human peripheral blood may be a potential indicator in the diagnosis of breast cancer. Oncol Res Treat. 2017;40:603–8.
Toor SM, Syed Khaja AS, El Salhat H, Bekdache O, Kanbar J, Jaloudi M, et al. Increased levels of circulating and tumor-infiltrating granulocytic myeloid cells in colorectal cancer patients. Front Immunol. 2016;7:560.
Weide B, Martens A, Zelba H, Stutz C, Derhovanessian E, Di Giacomo AM, et al. Myeloid-derived suppressor cells predict survival of patients with advanced melanoma: comparison with regulatory T cells and NY-ESO-1- or melan-A-specific T cells. Clin Cancer Res. 2014;20:1601–9.
Huang H, Zhang G, Li G, Ma H, Zhang X. Circulating CD14(+)HLA-DR(−/low) myeloid-derived suppressor cell is an indicator of poor prognosis in patients with ESCC. Tumour Biol. 2015;36:7987–96.
Angell TE, Lechner MG, Smith AM, Martin SE, Groshen SG, Maceri DR, et al. Circulating myeloid-derived suppressor cells predict differentiated thyroid cancer diagnosis and extent. Thyroid. 2016;26:381–9.
Tian T, Gu X, Zhang B, Liu Y, Yuan C, Shao L, et al. Increased circulating CD14(+)HLA-DR−/low myeloid-derived suppressor cells are associated with poor prognosis in patients with small-cell lung cancer. Cancer Biomark: Sect A Dis Markers. 2015;15:425–32.
Yang G, Shen W, Zhang Y, Liu M, Zhang L, Liu Q, et al. Accumulation of myeloid-derived suppressor cells (MDSCs) induced by low levels of IL-6 correlates with poor prognosis in bladder cancer. Oncotarget. 2017;8:38378–88.
Soler DC, Young AB, Cooper KD, Kerstetter-Fogle A, Barnholtz-Sloan JS, Gittleman H, et al. The ratio of HLA-DR and VNN2(+) expression on CD14(+) myeloid derived suppressor cells can distinguish glioblastoma from radiation necrosis patients. J Neurooncol. 2017;134:189–96.
Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19:108–19.
Pencovich N, Hantisteanu S, Wurtzel O, Hallak M, Fainaru O. Unique expression patterns associated with preferential recruitment of immature myeloid cells into angiogenic versus dormant tumors. Genes Immun. 2013;14:90–8.
Fainaru O, Pencovich N, Hantisteanu S, Yona G, Hallak M. Immature myeloid cells derived from mouse placentas and malignant tumors demonstrate similar proangiogenic transcriptional signatures. Fertil Steril. 2013;99:910–7 e2.
Nueda MJ, Tarazona S, Conesa A. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics. 2014;30:2598–602.
Wang D, An G, Xie S, Yao Y, Feng G. The clinical and prognostic significance of CD14(+)HLA-DR(−/low) myeloid-derived suppressor cells in hepatocellular carcinoma patients receiving radiotherapy. Tumour Biol. 2016;37:10427–33.
Arihara F, Mizukoshi E, Kitahara M, Takata Y, Arai K, Yamashita T, et al. Increase in CD14+HLA-DR−/low myeloid-derived suppressor cells in hepatocellular carcinoma patients and its impact on prognosis. Cancer Immunol Immunother. 2013;62:1421–30.
Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49–59.
Jordan KR, Amaria RN, Ramirez O, Callihan EB, Gao D, Borakove M, et al. Myeloid-derived suppressor cells are associated with disease progression and decreased overall survival in advanced-stage melanoma patients. Cancer Immunol Immunother. 2013;62:1711–22.
Zaslavsky A, Chen C, Grillo J, Baek KH, Holmgren L, Yoon SS, et al. Regional control of tumor growth. Mol Cancer Res. 2010;8:1198–206.
Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–74.
Giallongo C, Tibullo D, Parrinello NL, La Cava P, Di Rosa M, Bramanti V, et al. Granulocyte-like myeloid derived suppressor cells (G-MDSC) are increased in multiple myeloma and are driven by dysfunctional mesenchymal stem cells (MSC). Oncotarget. 2016;7:85764–75.
Hawila E, Razon H, Wildbaum G, Blattner C, Sapir Y, Shaked Y, et al. CCR5 directs the mobilization of CD11b(+)Gr1(+)Ly6C(low) polymorphonuclear myeloid cells from the bone marrow to the blood to support tumor development. Cell Rep. 2017;21:2212–22.
Pio R, Ajona D, Lambris JD. Complement inhibition in cancer therapy. Semin Immunol. 2013;25:54–64.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36.
Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–86.
Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10.
Acknowledgements
GF is the incumbent of the David and Stacey Cynamon Research fellow Chair in genetics and personalized medicine.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Nachmany, I., Bogoch, Y., Friedlander-Malik, G. et al. The transcriptional profile of circulating myeloid derived suppressor cells correlates with tumor development and progression in mouse. Genes Immun 20, 589–598 (2019). https://doi.org/10.1038/s41435-019-0062-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-019-0062-3
This article is cited by
-
CC-01 (chidamide plus celecoxib) modifies the tumor immune microenvironment and reduces tumor progression combined with immune checkpoint inhibitor
Scientific Reports (2022)
-
High-density neutrophils in MGUS and multiple myeloma are dysfunctional and immune-suppressive due to increased STAT3 downstream signaling
Scientific Reports (2020)