Abstract
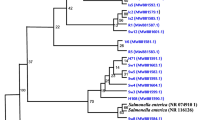
Escherichia coli (E. coli) are widely related to pyometra and cystitis in dogs, and these infections can occur simultaneously. The goal of this study was to determine genetic and pathogenic insights of 14 E. coli isolated simultaneously from pyometra content and bladder urine of seven bitches. To achieve this, in silico and in vitro comparative analyses were conducted. Whole-genome comparisons demonstrated that E. coli isolated from pyometra and urine of the same animal were predominantly genetic extraintestinal E. coli clones belonging to the same Sequence Type and phylogroup. The E. coli clones identified in this study included ST372, ST457, ST12, ST127, ST646, and ST961. Five isolates (35.7%) belonged to the ST12 complex. Except for two E. coli, all other isolates belonged to the B2 Clermont phylogroup. Interestingly, some genomes of E. coli from urine carried more virulence genes than those E. coli from pyometra. Both pyometra and urine E. coli isolates demonstrated a strong affinity for adhering to HeLa and T24 cells, with a low affinity for invading them. However, certain isolates from urine exhibited a greater tendency to adhere to T24 cells in qualitative and quantitative assays compared to isolates from pyometra. In conclusion, this study revealed the high genomic similarity between pyometra and urine E. coli isolates, as well as the virulent capacity of both to colonize endometrial and urothelial cells. The findings of this study underscore the importance of concurrently managing both infections clinically and could potentially contribute to future resources for the prevention of cystitis and pyometra.







Similar content being viewed by others
References
Alikhan NF, Petty NK, Zakour NL, Beatson SA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:1–10. https://doi.org/10.1186/1471-2164-12-402
Beghain J, Bridier-Nahmias A, Nagard H, Denamur E, Clermont O (2018) ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom 4(7). https://doi.org/10.1099/mgen.0.000192
Coggan JA, Melville PA, Oliveira CM, Faustino M, Moreno AM, Benites NR (2008) Microbiological and histopathological aspects of canine pyometra. Braz J Microbiol 39:477–483. https://doi.org/10.1590/S1517-838220080003000012
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:1–19. https://doi.org/10.1186/1471-2105-5-113
Edwards AM, Massey RC (2011) Invasion of human cells by a bacterial pathogen. J Vis Exp 49:e2693. https://doi.org/10.3791/2693
Elsinghorst EA (1994) Measurement of invasion by gentamicin resistance. Methods Enzymol 236:405–420. https://doi.org/10.1016/0076-6879(94)36030-8
Enterobase: Escherichia/ Shigella (2022). https://enterobase.warwick.ac.uk/species/ecoli/search_strains?query=st_search. Accessed 26 Oct 2022
Flament-Simon SC, de Toro M, García V, Blanco JE, Blanco M, Alonso MP, Goicoa A, Díaz-gonzález J, Nicolas-chanoine MH, Blanco J (2020) Molecular Characteristics of Extraintestinal Pathogenic E. coli (ExPEC), Uropathogenic E. coli (UPEC), and Multidrug Resistant E. coli Isolated from Healthy Dogs in Spain. Whole Genome Sequencing of Canine ST372 Isolates and Comparison with Human Isolates Causing Extraintestinal Infections. Microorganisms 8:1–25. https://doi.org/10.3390/microorganisms8111712
Gyles CL, Prescott JF, Songer G, Thoen CO (2010) Pathogenesis of bacterial infections in animals. Wiley, New Jersey
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29(8):1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Hagman R, Kühn I (2002) Escherichia coli strains isolated from the uterus and urinary bladder of bitches suffering from pyometra: comparison by restriction enzyme digestion and pulsed-field gel electrophoresis. Vet Microbiol 84:143–153. https://doi.org/10.1016/s0378-1135(01)00449-7
Henriques S, Silva E, Silva MF, Carvalho S, Diniz P, Lopes-Da-Costa L, Mateus L (2016) Immunomodulation in the canine endometrium by uteropathogenic Escherichia coli Vet Res 47:1–17. https://doi.org/10.1186/s13567-016-0396-z
Hernandez D, François P, Farinelli L, Østerås M, Schrenzel J (2008) De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res 18:802–809. https://doi.org/10.1101/gr.072033.107
Huang Y, Niu B, Gao Y, Fu L, Li W (2010) CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. https://doi.org/10.1093/bioinformatics/btq003
Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, Birol I (2017) ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res 27:768–777. https://doi.org/10.1101/gr.214346.116
Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F (2015) Rapid and Easy in Silico Serotyping of Escherichia coli isolates by Use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. https://doi.org/10.1128/JCM.00008-15
Jolley KA, Bray JE, Maiden MCJ (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:1–11. https://doi.org/10.1186/1471-2105-11-595
Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O (2014) Solving the Problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 9:e104984. https://doi.org/10.1371/journal.pone.0104984
Letourneau J, Levesque C, Berthiaume F, Jacques M, Mourez M (2011) In vitro assay of bacterial adhesion onto mammalian epithelial cells. J Vis Exp 51:e2783. https://doi.org/10.3791/2783
Lopes CE, de Carli S, Riboldi CI, de Lorenzo C, Panziera W, Driemeier D, Siqueira FM (2021) Pet Pyometra: correlating Bacteria pathogenicity to endometrial histological changes. Pathogens 10:833. https://doi.org/10.3390/pathogens10070833
Lopes CE, de Carli S, Weber MN, Fonseca ACV, Tagliari NJ, Foresti L, Cibulski SP, Mayer FQ, Canal CW, Siqueira FM (2020) Insights on the genetic features of endometrial pathogenic Escherichia coli strains from pyometra in companion animals: improving the knowledge about pathogenesis. Infect Genet Evol 85:104453. https://doi.org/10.1016/j.meegid.2020.104453
Mateus L, Henriques S, Merino C, Pomba C, Lopes da Costa L, Silva E (2013) Virulence genotypes of Escherichia coli canine isolates from pyometra, cystitis and fecal origin. Vet Microbiol 166:590–594. https://doi.org/10.1016/j.vetmic.2013.07.018
Melo RT, Oliveira RP, Silva BF, Monteiro GP, Saut JPE, Costa LRM, Dias SDC, Rossi DA (2022) Phylogeny and virulence factors of Escherichia coli isolated from dogs with Pyometra. Vet Sci 9:158. https://doi.org/10.3390/vetsci9040158
Mulvey MA, Schilling JD, Hultgren SJ (2001) Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69:4572–4579. https://doi.org/10.1128/IAI.69.7.4572-4579.2001
O’Brien VP, Gilbert NM, Lebratti T, Agarwal K, Foster L, Shin H, Lewis AL (2019) Low-dose inoculation of Escherichia coli achieves robust vaginal colonization and results in ascending infection accompanied by severe uterine inflammation in mice. PLoS ONE 14:e0219941. https://doi.org/10.1371/journal.pone.0219941
Robertson J, Nash JHE (2018) MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom 4:8. https://doi.org/10.1099/MGEN.0.000206
Roer L, Tchesnokova V, Allesøe R, Muradova M, Chattopadhyay S, Ahrenfeldt J, Thomsen MCF, Lund O, Hansen F, Hammerum AM, Sokurenko E, Hasman H (2017) Development of a web tool for Escherichia coli subtyping based on fimH alleles. J Clin Microbiol 55:2538–2543. https://doi.org/10.1128/JCM.00737-17
Siqueira FM, de Carli S, Lopes CE, Machado L, Vieira TR, Pöppl G, Cardoso MRI, Zaha A (2021) Non-lactose-fermenting uropathogenic Escherichia coli from dogs: virulence profile characterization. Lett Appl Microbiol 72:596–603. https://doi.org/10.1111/lam.13454
Siqueira AK, Ribeiro MG, Leite DS, Tiba MR, Moura C, Lopes MD, Prestes NC, Salerno T, Silva AV (2009) Virulence factors in Escherichia coli strains isolated from urinary tract infection and pyometra cases and from feces of healthy dogs. Res Vet Sci 86:206–210. https://doi.org/10.1016/j.rvsc.2008.07.018
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Tetzschner AMM, Johnson JR, Johnston BD, Lund O, Scheutz F (2020) In Silico Genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol 58:e01269-01220. https://doi.org/10.1128/JCM.01269-20
Valat C, Drapeau A, Beurlet S, Bachy V, Boulouis HJ, Pin R, Cazeau G, Madec JY, Haenni M (2020) Pathogenic Escherichia coli in Dogs reveals the predominance of ST372 and the Human-Associated ST73 extra-intestinal lineages. Front Microbiol 11:580. https://doi.org/10.3389/fmicb.2020.00580
Wadås B, Kühn I, Lagerstedt AS, Jonsson P (1996) Biochemical phenotypes of Escherichia coli in dogs: comparison of isolates isolated from bitches suffering from pyometra and urinary tract infection with isolates from faeces of healthy dogs. Vet Microbiol 52:293–300. https://doi.org/10.1016/s0378-1135(96)00067-3
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542. https://doi.org/10.1093/nar/gkw1017
Xavier RGC, da Silva PHS, Trindade HD, Carvalho GM, Nicolino RR, Freitas PMC, Silva ROS (2022) Characterization of Escherichia coli in Dogs with Pyometra and the influence of Diet on the intestinal colonization of Extraintestinal Pathogenic E. Coli (ExPEC). Vet Sci 9:245. https://doi.org/10.3390/vetsci9050245
Acknowledgements
We thank Rede de Ciências Ômicas (RECOM), Dr. Claudio Wageck Canal, Dr. Ana Maria Oliveira Battastini, Dr. Annelise Kopp Alves, and Dr. Carlos Pérez Bergmann for the research collaboration. We also thank Lais Foresti for the support in sample collection and the Laboratory of Veterinary Bacteriology of the UFRGS team for technical support.
Funding
This work was supported by the Coordination of Improvement of Higher Educational Personnel (CAPES) [grant number 001].
Author information
Authors and Affiliations
Contributions
Conceptualization: Cassiane Elisabete Lopes, Tania Aparecida Tardelli Gomes, and Franciele Maboni Siqueira; Methodology: Cassiane Elisabete Lopes, Tania Aparecida Tardelli Gomes, Mateus Matiuzzi da Costa, Rommel Thiago Jucá Ramos, Aristóteles Góes Neto, Vasco Ariston de Carvalho Azevedo, Bertram Brenig, and Franciele Maboni Siqueira; Formal analysis and investigation: Cassiane Elisabete Lopes, Tainara Soares Weyh, Camila Azevedo Moni, Maria Eduarda Dias, and Franciele Maboni Siqueira; Writing - original draft preparation: Cassiane Elisabete Lopes; Writing - review and editing: Franciele Maboni Siqueira; Resources: Mateus Matiuzzi da Costa, Rommel Thiago Jucá Ramos, Aristóteles Góes Neto, Vasco Ariston de Carvalho Azevedo, Bertram Brenig; Supervision: Franciele Maboni Siqueira. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was performed in line with the principles of the Ethics Committee of the Federal University of Rio Grande do Sul (Ethics Committee number 31874).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
ESM 1
Online Resource 2. Circular comparison among Escherichia coli genomes. A BLAST alignment was performed with the 14 genomes of this study and one characterized EnPEC genome (E. coli LBV005/17 - QLNJ00000000) against the reference UPEC genome (E. coli CFT 073 - AE014075.1) using the BRIG software. The image demonstrates a high similarity of the E. coli isolates with some punctual polymorphisms among them. Online Resource 3. Matrix of SNP differences among the Escherichia coli genomes of the study. The SNPs distance matrix was constructed by CSI Phylogeny v.1.4. The number of SNPs differences among the 14 assembled genomes of this study was demonstrated on each matrix box. A color scale was added to the matrix, in which the darker the green, the lower the number of SNPs among the genomes. E. coli CFT 073 (AE014075.1) and E. coli LBV005/17 (QLNJ00000000) were used as UPEC and EnPEC reference control genomes, respectively. Online Resource 4. Adhesion pattern of a set of pyometra and urine Escherichia coli isolates on HeLa cells by optical microscopy. The adherence pattern of the randomly chosen isolates E. coli LBV064/17_pyometra (A) and E. coli LBV064/17_urine (B) was obtained after a 3 h adhesion assay with HeLa cells. Light microscopy images (400X microscopic magnification) show bacterial clumps with affinity to HeLa cells (arrows).E. coli CFT 073 as positive control (C). E. coli EAEC 042 shows typical aggregative adhesion (D). Non-infected HeLa cells (E). Online Resource 5. Adhesion pattern of a set of pyometra and urine Escherichia coli isolates on T24 cells by optical microscopy. The adherence pattern of the randomly chosen isolatesE. coli LBV064/17_pyometra (A) and E. coli LBV064/17_urine (B) was obtained after a 3 h adhesion assay with the T24 cell line. E. coli UPEC CFT 073 as positive control (C). Light microscopy images (400X microscopic magnification) show affinity with the formation of bacterial clumps on T24 cells (arrows). E. coli EAEC 042 shows typical aggregative adhesion (D). Non-infected T24 cells (200X microscopic magnification) (E). Online Resource 6. Electron microscopy evaluation of a set of Escherichia coli isolates adhesion on HeLa cells compared to control strains. Electron microscopy images (3,300X microscopic magnification) were obtained after adhesion assays with 3 h of incubation. The adherence pattern of the randomly selected isolates E. coli LBV064/17_pyometra (A) and E. coli LBV064/17_urine (B) demonstrated diffuse adhesion with some grouped bacteria on HeLa surface. This adhesion pattern was very similar to the UPEC control strain CFT 073 (C). E. coli EAEC 042 exhibited typical aggregative adhesion (D). Online Resource 7. Electron microscopy evaluation of a set of Escherichia coli isolates adhesion on T24 cells compared to control strains. Electron microscopy images (3,300X microscopic magnification) were obtained after an adhesion assay with 3 h of incubation. The adherence pattern of the randomly selected isolates E. coli LBV064/17_pyometra (A) and E. coli LBV064/17_urine (B). Suggestive biofilm formation of E. coli LBV064/17_urine was frequently identified on the surface of T24 cells (arrows). The biofilm production was less frequent on the E. coli LBV064/17_pyometra sample. E. coli CFT 073 exhibited a high level of affinity to T24 cells (C). E. coli. EAEC 042 showcased typical aggregative adhesion (D) Online Resource 10. Virulence genes profile of the Escherichia coli isolated from bitch pyometra and urine. The graphic represents the frequency of each gene or family of genes found in the 14 E. coli genomes of this study. The genes were grouped by different pathogenic tools of E. coli (cytotoxins, microcins, biofilm production, host immune modulation, polysaccharide capsule production, and iron uptake systems). The search for genomic virulence genes was performed by PATRIC and Virulence Finder databases. (ZIP 358 MB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lopes, C.E., Weyh, T.S., Moni, C.A. et al. Comparative genetic and pathogenic approaches of Escherichia coli isolated simultaneously from pyometra and urine of bitches. Vet Res Commun (2024). https://doi.org/10.1007/s11259-024-10355-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11259-024-10355-7