“It’s Only a Model”: When Protein Structure Predictions Need Experimental Validation, the Case of the HTLV-1 Tax Protein
Abstract
:1. Introduction
2. Predicting the Structure of HTLV-1 Tax
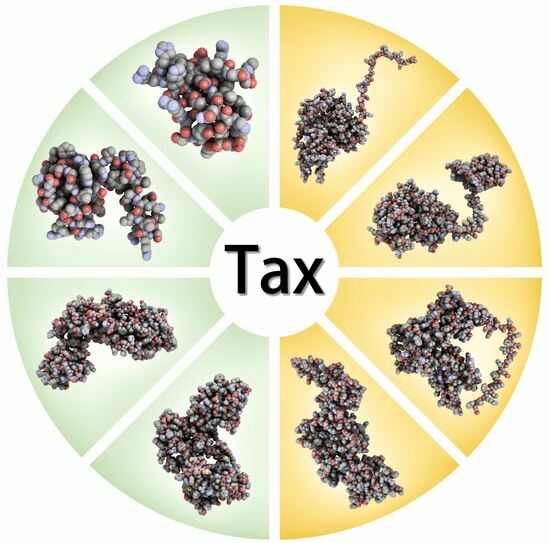
2.1. Predictions Using Homology Modelling
2.2. Predictions Using AI-Based Pipeline
3. Comparison of HTLV-1 Tax Structure Models
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poiesz, B.J.; Ruscetti, F.W.; Gazdar, A.F.; Bunn, P.A.; Minna, J.D.; Gallo, R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 1980, 77, 7415–7419. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Cassar, O. Epidemiological aspects and world distribution of HTLV-1 infection. Front. Microbiol. 2012, 3, 388. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.P.; Al-Saleem, J.; Green, P.L. Comparative virology of HTLV-1 and HTLV-2. Retrovirology 2019, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Currer, R.; van Duyne, R.; Jaworski, E.; Guendel, I.; Sampey, G.; Das, R.; Narayanan, A.; Kashanchi, F. HTLV tax: A fascinating multifunctional co-regulator of viral and cellular pathways. Front. Microbiol. 2012, 3, 406. [Google Scholar] [CrossRef] [PubMed]
- Boxus, M.; Twizere, J.-C.; Legros, S.; Dewulf, J.-F.; Kettmann, R.; Willems, L. The HTLV-1 Tax interactome. Retrovirology 2008, 5, 76. [Google Scholar] [CrossRef] [PubMed]
- Martella, C.; Waast, L.; Pique, C. Tax, marionnettiste de la transcription du HTLV-1. Med. Sci. 2022, 38, 359–365. [Google Scholar] [CrossRef]
- Fochi, S.; Mutascio, S.; Bertazzoni, U.; Zipeto, D.; Romanelli, M.G. HTLV deregulation of the NF-kappaB pathway: An update on Tax and antisense proteins role. Front. Microbiol. 2018, 9, 285. [Google Scholar] [CrossRef]
- Mohanty, S.; Harhaj, E.W. Mechanisms of Oncogenesis by HTLV-1 Tax. Pathogens 2020, 9, 543. [Google Scholar] [CrossRef]
- Simonis, N.; Rual, J.-F.; Lemmens, I.; Boxus, M.; Hirozane-Kihijawa, T.; Gatot, J.-S.; Dricot, A.; Hao, T.; Vertommen, D.; Legros, S.; et al. Host-pathogen interactome mapping for HTLV-1 and HTLV-2. Retrovirology 2012, 9, 26. [Google Scholar] [CrossRef]
- Fiorini, F.; Robin, J.P.; Kanaan, J.; Borowiak, M.; Croquette, V.; Le Hir, H.; Jalinot, P.; Mocquet, V. HTLV-1 Tax plugs and freezes UPF1 helicase leading to nonsense-mediated mRNA decay inhibition. Nat. Commun. 2018, 9, 431. [Google Scholar] [CrossRef]
- Higuchi, M.; Fujii, M. Distinct functions of HTLV-1 Tax1 from HTLV-2 Tax2 contribute key roles to viral pathogenesis. Retrovirology 2009, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Hleihel, R.; Skayneh, H.; de The, H.; Hermine, O.; Bazarbachi, A. Primary cells from patients with adult T cell leukemia/lymphoma depend on HTLV-1 Tax expression for NF-kappaB activation and survival. Blood Cancer J. 2023, 13, 67. [Google Scholar] [CrossRef]
- Garboczi, D.N.; Ghosh, P.; Utz, U.; Fan, Q.R.; Biddison, W.E.; Wiley, D.C. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 1996, 384, 134–141. [Google Scholar] [CrossRef]
- Gagnon, S.J.; Borbulevych, O.Y.; Davis-Harrison, R.L.; Baxter, T.K.; Clemens, J.R.; Armstrong, K.M.; Turner, R.V.; Damirjian, M.; Biddison, W.E.; Baker, B.M. Unraveling a hotspot for TCR recognition on HLA-A2: Evidence against the existence of peptide-independent TCR binding determinants. J. Mol. Biol. 2005, 353, 556–573. [Google Scholar] [CrossRef]
- Ding, Y.H.; Smith, K.J.; Garboczi, D.N.; Utz, U.; Biddison, W.E.; Wiley, D.C. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity 1998, 8, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.H.; Baker, B.M.; Garboczi, D.N.; Biddison, W.E.; Wiley, D.C. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity 1999, 11, 45–56. [Google Scholar] [CrossRef]
- Gagnon, S.J.; Borbulevych, O.Y.; Davis-Harrison, R.L.; Turner, R.V.; Damirjian, M.; Wojnarowicz, A.; Biddison, W.E.; Baker, B.M. T cell receptor recognition via cooperative conformational plasticity. J. Mol. Biol. 2006, 363, 228–243. [Google Scholar] [CrossRef]
- Piepenbrink, K.H.; Borbulevych, O.Y.; Sommese, R.F.; Clemens, J.; Armstrong, K.M.; Desmond, C.; Do, P.; Baker, B.M. Fluorine substitutions in an antigenic peptide selectively modulate T-cell receptor binding in a minimally perturbing manner. Biochem. J. 2009, 423, 353–361. [Google Scholar] [CrossRef]
- Khan, A.R.; Baker, B.M.; Ghosh, P.; Biddison, W.E.; Wiley, D.C. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J. Immunol. 2000, 164, 6398–6405. [Google Scholar] [CrossRef]
- Singh, N.K.; Alonso, J.A.; Harris, D.T.; Anderson, S.D.; Ma, J.; Hellman, L.M.; Rosenberg, A.M.; Kolawole, E.M.; Evavold, B.D.; Kranz, D.M.; et al. An Engineered T Cell Receptor Variant Realizes the Limits of Functional Binding Modes. Biochemistry 2020, 59, 4163–4175. [Google Scholar] [CrossRef] [PubMed]
- Borbulevych, O.Y.; Piepenbrink, K.H.; Gloor, B.E.; Scott, D.R.; Sommese, R.F.; Cole, D.K.; Sewell, A.K.; Baker, B.M. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity 2009, 31, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Muzahim, Y.; Bouvier, M. Crystal structure of adenovirus E3-19K bound to HLA-A2 reveals mechanism for immunomodulation. Nat. Struct. Mol. Biol. 2012, 19, 1176–1181. [Google Scholar] [CrossRef]
- Madden, D.R.; Garboczi, D.N.; Wiley, D.C. The antigenic identity of peptide-MHC complexes: A comparison of the conformations of five viral peptides presented by HLA-A2. Cell 1993, 75, 693–708. [Google Scholar] [CrossRef]
- Gogl, G.; Zambo, B.; Kostmann, C.; Cousido-Siah, A.; Morlet, B.; Durbesson, F.; Negroni, L.; Eberling, P.; Jane, P.; Nomine, Y.; et al. Quantitative fragmentomics allow affinity mapping of interactomes. Nat. Commun. 2022, 13, 5472. [Google Scholar] [CrossRef]
- Maseko, S.B.; Brammerloo, Y.; van Molle, I.; Sogues, A.; Martin, C.; Gorgulla, C.; Plant, E.; Olivet, J.; Blavier, J.; Ntombela, T.; et al. Identification of small molecule antivirals against HTLV-1 by targeting the hDLG1-Tax-1 protein-protein interaction. Antiviral Res. 2023, 217, 105675. [Google Scholar] [CrossRef]
- Cousido-Siah, A.; Carneiro, L.; Kostmann, C.; Ecsedi, P.; Nyitray, L.; Trave, G.; Gogl, G. A scalable strategy to solve structures of PDZ domains and their complexes. Acta Crystallogr. D Struct. Biol. 2022, 78, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Javorsky, A.; Maddumage, J.C.; Mackie, E.R.R.; Soares da Costa, T.P.; Humbert, P.O.; Kvansakul, M. Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 PBM. FEBS J. 2023, 290, 974–987. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Kuzin, A.P.; Abashidze, M.; Seetharaman, J.; Vorobiev, S.M.; Wang, D.; Fang, Y.; Owens, L.; Ma, L.-C.; Xiao, R.; Liu, J.; et al. Crystal Structure of the Putative Nitrite Reductase NADPH (Small Subunit) Oxidoreductase Protein Q87HB1. Available online: https://www.wwpdb.org/pdb?id=pdb_00003c0d (accessed on 3 February 2024).
- Moe, L.A.; Bingman, C.A.; Wesenberg, G.E.; Phillips, G.N., Jr.; Fox, B.G. Structure of T4moC, the Rieske-type ferredoxin component of toluene 4-monooxygenase. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Bruns, C.M.; Karplus, P.A. Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: Oxidized, reduced and 2′-phospho-5′-AMP bound states. J. Mol. Biol. 1995, 247, 125–145. [Google Scholar] [CrossRef]
- Raza, M.T.; Mizan, S.; Yasmin, F.; Akash, A.S.; Shahik, S.M. Epitope-based universal vaccine for Human T-lymphotropic virus-1 (HTLV-1). PLoS ONE 2021, 16, e0248001. [Google Scholar] [CrossRef]
- Schulman, B.A.; Carrano, A.C.; Jeffrey, P.D.; Bowen, Z.; Kinnucan, E.R.; Finnin, M.S.; Elledge, S.J.; Harper, J.W.; Pagano, M.; Pavletich, N.P. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 2000, 408, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Toesca, J.; Guillon, C. Review and perspectives on the structure-function relationships of the Gag subunits of Feline Immunodeficiency Virus. Pathogens 2021, 10, 1502. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef]
- Zheng, W.; Wuyun, Q.; Freddolino, P.L.; Zhang, Y. Integrating deep learning, threading alignments, and a multi-MSA strategy for high-quality protein monomer and complex structure prediction in CASP15. Proteins 2023, 91, 1684–1703. [Google Scholar] [CrossRef]
- Yuan, S.; Yu, X.; Topf, M.; Dorstyn, L.; Kumar, S.; Ludtke, S.J.; Akey, C.W. Structure of the Drosophila apoptosome at 6.9 a resolution. Structure 2011, 19, 128–140. [Google Scholar] [CrossRef]
- Kluska, K.; Adamczyk, J.; Krezel, A. Metal binding properties of zinc fingers with a naturally altered metal binding site. Metallomics 2018, 10, 248–263. [Google Scholar] [CrossRef]
- Hajikhezri, Z.; Darweesh, M.; Akusjarvi, G.; Punga, T. Role of CCCH-Type Zinc Finger Proteins in Human Adenovirus Infections. Viruses 2020, 12, 1322. [Google Scholar] [CrossRef]
- Yuan, S.; Xu, B.; Zhang, J.; Xie, Z.; Cheng, Q.; Yang, Z.; Cai, Q.; Huang, B. Comprehensive analysis of CCCH-type zinc finger family genes facilitates functional gene discovery and reflects recent allopolyploidization event in tetraploid switchgrass. BMC Genom. 2015, 16, 129. [Google Scholar] [CrossRef]
- Foucault, M.; Mayol, K.; Receveur-Bréchot, V.; Bussat, M.-C.; Klinguer-Hamour, C.; Verrier, B.; Beck, A.; Haser, R.; Gouet, P.; Guillon, C. UV and X-ray structural studies of a 101-residue long Tat protein from a HIV-1 primary isolate and of its mutated, detoxified, vaccine candidate. Proteins 2010, 78, 1441–1456. [Google Scholar] [CrossRef] [PubMed]
- Shojania, S.; O’Neil, J.D. HIV-1 Tat is a natively unfolded protein: The solution conformation and dynamics of reduced HIV-1 Tat-(1-72) by NMR spectroscopy. J. Biol. Chem. 2006, 281, 8347–8356. [Google Scholar] [CrossRef] [PubMed]
- Tahirov, T.H.; Babayeva, N.D.; Varzavand, K.; Cooper, J.J.; Sedore, S.C.; Price, D.H. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 2010, 465, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Bex, F.; Murphy, K.; Wattiez, R.; Burny, A.; Gaynor, R.B. Phosphorylation of the human T-cell leukemia virus type 1 transactivator tax on adjacent serine residues is critical for tax activation. J. Virol. 1999, 73, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Lodewick, J.; Lamsoul, I.; Polania, A.; Lebrun, S.; Burny, A.; Ratner, L.; Bex, F. Acetylation of the human T-cell leukemia virus type 1 Tax oncoprotein by p300 promotes activation of the NF-kappaB pathway. Virology 2009, 386, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Lavorgna, A.; Harhaj, E.W. Regulation of HTLV-1 tax stability, cellular trafficking and NF-kappaB activation by the ubiquitin-proteasome pathway. Viruses 2014, 6, 3925–3943. [Google Scholar] [CrossRef]
- Steinacher, R.; Schar, P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 2005, 15, 616–623. [Google Scholar] [CrossRef]
- Kumar, A.; Narayanan, V.; Sekhar, A. Characterizing Post-Translational Modifications and Their Effects on Protein Conformation Using NMR Spectroscopy. Biochemistry 2020, 59, 57–73. [Google Scholar] [CrossRef]
- Philipsen, L.; Reddycherla, A.V.; Hartig, R.; Gumz, J.; Kastle, M.; Kritikos, A.; Poltorak, M.P.; Prokazov, Y.; Turbin, E.; Weber, A.; et al. De novo phosphorylation and conformational opening of the tyrosine kinase Lck act in concert to initiate T cell receptor signaling. Sci. Signal 2017, 10, eaaf4736. [Google Scholar] [CrossRef]
- Gres, A.T.; Kirby, K.A.; KewalRamani, V.N.; Tanner, J.J.; Pornillos, O.; Sarafianos, S.G. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 2015, 349, 99–103. [Google Scholar] [CrossRef]
- Kwong, P.D.; Wyatt, R.; Majeed, S.; Robinson, J.; Sweet, R.W.; Sodroski, J.; Hendrickson, W.A. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold. Des. 2000, 8, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, L.; Schwieters, C.D.; Grishaev, A.; Clore, G.M. Quantitative Characterization of Configurational Space Sampled by HIV-1 Nucleocapsid Using Solution NMR, X-ray Scattering and Protein Engineering. Chemphyschem 2016, 17, 1548–1552. [Google Scholar] [CrossRef]
- Chakravarty, D.; Porter, L.L. AlphaFold2 fails to predict protein fold switching. Protein Sci. 2022, 31, e4353. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Liebschner, D.; Croll, T.I.; Williams, C.J.; McCoy, A.J.; Poon, B.K.; Afonine, P.V.; Oeffner, R.D.; Richardson, J.S.; Read, R.J.; et al. AlphaFold predictions are valuable hypotheses and accelerate but do not replace experimental structure determination. Nat. Methods 2024, 21, 110–116. [Google Scholar] [CrossRef] [PubMed]






| Modelling Server | Complete Protein Modelled? | Calculated QMEANDisCo |
|---|---|---|
| Swiss-Model | No | 0.31 ± 0.12 |
| Phyre2 (default) | No | 0.35 ± 0.11 |
| Phyre2 (intensive) | Yes | 0.27 ± 0.05 |
| I-Tasser | Yes | 0.35 ± 0.05 |
| Modelling Program | Complete Protein Modelled? | Original Confidence Score | Calculated QMEANDisCo |
|---|---|---|---|
| AlphaFold 2 | Yes | pLDDT = 37.4 | 0.35 ± 0.05 |
| RoseTTAFold | Yes | Predicted GDT = 0.35 | 0.39 ± 0.05 |
| ESMFold | Yes | pLDDT = 47.6 | 0.43 ± 0.05 |
| D-I-Tasser | Yes | eTM = 0.4 | 0.44 ± 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guillon, C.; Robert, X.; Gouet, P. “It’s Only a Model”: When Protein Structure Predictions Need Experimental Validation, the Case of the HTLV-1 Tax Protein. Pathogens 2024, 13, 241. https://doi.org/10.3390/pathogens13030241
Guillon C, Robert X, Gouet P. “It’s Only a Model”: When Protein Structure Predictions Need Experimental Validation, the Case of the HTLV-1 Tax Protein. Pathogens. 2024; 13(3):241. https://doi.org/10.3390/pathogens13030241
Chicago/Turabian StyleGuillon, Christophe, Xavier Robert, and Patrice Gouet. 2024. "“It’s Only a Model”: When Protein Structure Predictions Need Experimental Validation, the Case of the HTLV-1 Tax Protein" Pathogens 13, no. 3: 241. https://doi.org/10.3390/pathogens13030241