Abstract
Astrocytes, the most abundant glial cell type in the brain, are underrepresented in traditional cortical organoid models due to the delayed onset of cortical gliogenesis. Here we introduce a new glia-enriched cortical organoid model that exhibits accelerated astrogliogenesis. We demonstrated that induction of a gliogenic switch in a subset of progenitors enabled the rapid derivation of astroglial cells, which account for 25–31% of the cell population within 8–10 weeks of differentiation. Intracerebral transplantation of these organoids reliably generated a diverse repertoire of cortical neurons and anatomical subclasses of human astrocytes. Spatial transcriptome profiling identified layer-specific expression patterns among distinct subclasses of astrocytes within organoid transplants. Using an in vivo acute neuroinflammation model, we identified a subpopulation of astrocytes that rapidly activates pro-inflammatory pathways upon cytokine stimulation. Additionally, we demonstrated that CD38 signaling has a crucial role in mediating metabolic and mitochondrial stress in reactive astrocytes. This model provides a robust platform for investigating human astrocyte function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
snRNA-seq data are available in Gene Expression Omnibus (GEO) under the accession GSE185472. The following public datasets were used for snRNA-seq analysis: Allen Brain Institute human adult snRNA-seq data from multiple cortical areas (https://portal.brain-map.org/atlases-and-data/rnaseq/human-multiple-cortical-areas-smart-seq; accessed October 2022), snRNA-seq data from broad temporal coverage from fetal to adulthood stages of the Brodmann area 8, 9, 10 and 46 prefrontal cortex regions (GEO accession GSE168408) and snRNA-seq from 8-month-old cortical organoid transplants (GEO accession GSE190815). For single-nucleus analysis, we used hg19 human reference genome v1.2.0 and mm10 mouse reference genome v1.2.0 provided by 10x Genomics. The sequences and gene files used to build the references can be achieved at ftp://ftp.ensembl.org/pub/grch37/release-84/fasta/homo_sapiens/dna/ and ftp://ftp.ensembl.org/pub/grch37/release-84/gtf/homo_sapiens/ (for human hg19 genome); ftp://ftp.ensembl.org/pub/release-84/fasta/mus_musculus/dna/ and ftp://ftp.ensembl.org/pub/release-84/gtf/mus_musculus/ (for mouse mm10 genome). All other raw data used for plotting in the figures are provided as source data. Source data are provided with this paper.
References
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Qian, X., Song, H. & Ming, G. L. Brain organoids: advances, applications and challenges. Development 146, dev166074 (2019).
Wang, M., Zhang, L. & Gage, F. H. Modeling neuropsychiatric disorders using human induced pluripotent stem cells. Protein Cell 11, 45–59 (2020).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Kim, J., Koo, B. K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
Velasco, S. et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527 (2019).
Sloan, S. A. et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95, 779–790 (2017).
Qian, X. et al. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell 26, 766–781 (2020).
Molofsky, A. V. et al. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 26, 891–907 (2012).
Tchieu, J. et al. NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat. Biotechnol. 37, 267–275 (2019).
Canals, I. et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat. Methods 15, 693–696 (2018).
Santos, R. et al. Differentiation of inflammation-responsive astrocytes from glial progenitors generated from human induced pluripotent stem cells. Stem Cell Reports 8, 1757–1769 (2017).
Barbar, L. et al. CD49f is a novel marker of functional and reactive human iPSC-derived astrocytes. Neuron 107, 436–453 (2020).
Zhang, J. & Liu, Q. Cholesterol metabolism and homeostasis in the brain. Protein Cell 6, 254–264 (2015).
Daneman, R. & Prat, A. The blood-brain barrier. Cold Spring Harb. Perspect. Biol. 7, a020412 (2015).
Mansour, A. A. et al. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 36, 432–441 (2018).
Bao, Z. et al. Human cerebral organoid implantation alleviated the neurological deficits of traumatic brain injury in mice. Oxid. Med. Cell. Longev. 2021, 6338722 (2021).
Daviaud, N., Friedel, R. H. & Zou, H. Vascularization and engraftment of transplanted human cerebral organoids in mouse cortex. eNeuro 5, ENEURO.0219-18.2018 (2018).
Kitahara, T. et al. Axonal extensions along corticospinal tracts from transplanted human cerebral organoids. Stem Cell Reports 15, 467–481 (2020).
Shi, Y. et al. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 18, e3000705 (2020).
Revah, O. et al. Maturation and circuit integration of transplanted human cortical organoids. Nature 610, 319–326 (2022).
Jgamadze, D. et al. Structural and functional integration of human forebrain organoids with the injured adult rat visual system. Cell Stem Cell 30, 137–152 (2023).
Qian, X. et al. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 13, 565–580 (2018).
Qian, X. et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Morabito, S. et al. Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat. Genet. 53, 1143–1155 (2021).
Allen, N. J. & Eroglu, C. Cell biology of astrocyte-synapse interactions. Neuron 96, 697–708 (2017).
Stogsdill, J. A. et al. Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197 (2017).
Oberheim, N. A. et al. Uniquely hominid features of adult human astrocytes. J. Neurosci. 29, 3276–3287 (2009).
Zhang, Y. et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016).
Oberheim, N. A., Wang, X., Goldman, S. & Nedergaard, M. Astrocytic complexity distinguishes the human brain. Trends Neurosci. 29, 547–553 (2006).
Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020).
Li, Y. et al. Direct labeling and visualization of blood vessels with lipophilic carbocyanine dye DiI. Nat. Protoc. 3, 1703–1708 (2008).
Herring, C. A. et al. Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution. Cell 185, 4428–4447 (2022).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Sofroniew, M. V. & Vinters, H. V. Astrocytes: biology and pathology. Acta Neuropathol. 119, 7–35 (2010).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410 (2012).
Anderson, M. A. et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature 532, 195–200 (2016).
Tarrago, M. G. et al. A potent and specific CD38 inhibitor ameliorates age-related metabolic dysfunction by reversing tissue NAD+ decline. Cell Metab. 27, 1081–1095 e1010 (2018).
Sprenger, H. G. & Langer, T. The good and the bad of mitochondrial breakups. Trends Cell Biol. 29, 888–900 (2019).
Krencik, R. & Zhang, S. C. Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat. Protoc. 6, 1710–1717 (2011).
Palm, T. et al. Rapid and robust generation of long-term self-renewing human neural stem cells with the ability to generate mature astroglia. Sci. Rep. 5, 16321 (2015).
Han, X. et al. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell 12, 342–353 (2013).
Windrem, M. S. et al. A competitive advantage by neonatally engrafted human glial progenitors yields mice whose brains are chimeric for human glia. J. Neurosci. 34, 16153–16161 (2014).
Mariani, J. N., Zou, L. & Goldman, S. A. Human glial chimeric mice to define the role of glial pathology in human disease. Methods Mol. Biol. 1936, 311–331 (2019).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Bayraktar, O. A. et al. Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat. Neurosci. 23, 500–509 (2020).
Hodge, R. D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019).
Jorstad, N. L. et al. Transcriptomic cytoarchitecture reveals principles of human neocortex organization. Science 382, eadf6812 (2023).
Burda, J. E. et al. Divergent transcriptional regulation of astrocyte reactivity across disorders. Nature 606, 557–564 (2022).
Cowan, C. A. et al. Derivation of embryonic stem-cell lines from human blastocysts. New Engl. J. Med. 350, 1353–1356 (2004).
Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Mertens, J. et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 527, 95–99 (2015).
Goncalves, J. T. et al. In vivo imaging of dendritic pruning in dentate granule cells. Nat. Neurosci. 19, 788–791 (2016).
Longair, M. H., Baker, D. A. & Armstrong, J. D. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453–2454 (2011).
Kuwajima, M., Mendenhall, J. M. & Harris, K. M. Large-volume reconstruction of brain tissue from high-resolution serial section images acquired by SEM-based scanning transmission electron microscopy. Methods Mol. Biol. 950, 253–273 (2013).
Deerinck, T. J., Bushong, E. A., Thor, A. & Ellisman, M. H. NCMIR methods for 3D EM: a new protocol for preparation of biological specimens for serial block face scanning electron microscopy. Microscopy 1, 6–8 (2010).
Horstmann, H., Korber, C., Satzler, K., Aydin, D. & Kuner, T. Serial section scanning electron microscopy (S3EM) on silicon wafers for ultra-structural volume imaging of cells and tissues. PLoS ONE 7, e35172 (2012).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z. & Zhang, B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205 (2019).
Acknowledgements
We thank A. Mansour (Salk Institute) for providing the pCSC-CAG-GFP lentiviral labeled H9 ESC line, N. Hah for technical assistance with snRNA-seq and R. Garg for assistance with EM segmentation. We also thank M.L. Gage for editorial comments and A. Cao and T. Bartol for their help with 3DEM visualization. This work was supported by the American Heart Association and the Paul G. Allen Frontiers Group Grant (19PABHI34610000/TEAM LEADER: Fred H. Gage/2019), JPB Foundation, Annette C. Merle-Smith, Lynn and Edward Streim, the Milky Way Foundation, the Ray and Dagmar Dolby Family Fund and NIH (R37 AG072502-03, P30 AG062429-05, P30 AG068635-04, R01 AG070154-04, AG056306-07 and P01 AG051449-08). This work was also supported by the NGS Core Facility, the GT3 Core Facility, the Razavi Newman Integrative Genomics and Bioinformatics Core Facility and the Waitt Advanced Biophotonics Core Facility of the Salk Institute with funding from NIH–NCI (CCSG, P30 014195), the Chapman Foundation, NINDS R24 Core Grant, NEI and the Waitt Foundation. M.W. is supported by a Young Investigator Grant from the Brain & Behavior Research Foundation (BBRFNARSAD) and a Pioneer Fund Postdoctoral Scholar Award from the Salk Institute. Figures 2a, 2h, 4a, 5a, 6a and 6d were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
M.W., L.Z. and F.H.G. conceived the study and wrote the paper. M.W., L.Z., C.K.L. and S.F. performed cell culture and organoid differentiation. M.W., L.Z., C.K.L. and I.S.G. performed surgeries. S.L.P. assisted in surgeries. M.W. and I.S.G. performed snRNA-seq and scRNA-seq. M.W., C.Q. and E.M. performed NanoString GeoMx DSP. M.W., M.N.S., J.Y. and A.E.W. performed bioinformatics analyses. M.W. and L.Z. performed imaging analysis with assistance from L.L.X., C.K.L., S.C. and M.D.S. L.Z., S.W.N. and L.R.A. performed sample processing for SEM. S.W.N. and L.R.A. performed electron microscopic image analysis under the supervision of M.W., L.Z. and U.M. F.H.G. provided funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Rapid astrogliogenesis in glia-enriched cortical organoids.
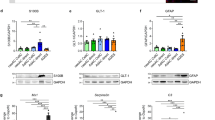
a. Sample bright-field images of hiPSCs/hESCs colonies and glia-enriched cortical organoids at days 0, 14, 30 and 60. Day 0 refers to embryoid bodies (EB). Scale bars, 890 μm. b. Quantitative PCR analysis of the expression levels of NFIA and SOX2 in three-week-old organoids cultured under varying conditions (Hues6: 1, n = 5; 2, n = 5; 3, n = 5; 4, n = 5; 5, n = 4. iPSC822: 1, n = 5; 2, n = 5; 3, n = 5; 4, n = 5; 5, n = 4). Each dot represents a pool of three organoids. Bars, mean ± s.d. Two-sided t-test, ns, not significant, *p < 0.05, ***p < 0.001, ****p < 0.0001. c. Immunostaining of two-month-old glia-enriched cortical organoids: stem cells (SOX2, magenta), astrocytes (GFAP, green). Scale bar, 100 μm. d. Immunostaining of two-month-old glia-enriched cortical organoids: intermediate progenitor cells (EOMES, green), cortical excitatory neurons (CTIP2, magenta; SATB2, gray), stem cells (SOX2, green), neurons (NeuN, magenta), astrocytes (GFAP, gray), glia (HOPX, green; S100B, magenta; GFAP, gray). Scale bars, 20 μm. e. Immunostaining of three-month-old glia-enriched cortical organoids differentiated in 2% FBS (top left panel), 2% SATO (top right panel; serum-free condition, SATO component reported in ref. 30), 10% FBS (bottom left panel) and 10% SATO (bottom right panel). Astrocytes (GFAP, green). Scale bars, 100 μm. f. Quantification of the GFAP fluorescence intensity in glia-enriched cortical organoids differentiated in 2% FBS, 2% SATO, 10% FBS and 10% SATO conditions (Hues6: 10% FBS, n = 3; 10% SATO, n = 6; 2% FBS, n = 4; 2% SATO, n = 6. iPSC822: 10% FBS, n = 6; 10% SATO, n = 8; 2% FBS, n = 5; 2% SATO, n = 6). Each dot represents one organoid. Bars, mean ± s.e.m. Two-sided t-test, ***p < 0.001, ****p < 0.0001.
Extended Data Fig. 2 snRNA-seq of 10-week-old glia-enriched cortical organoids.
a. UMAP plot of snRNA-seq data from 10-week-old glia-enriched cortical organoids. The percentage of each major cell types is shown. APC, astrocyte progenitor cell; Ast, astrocyte; IPC, intermediate progenitor cell; In, inhibitory neuron; Cortical Ex, cortical excitatory neuron; Ex1 and Ex2, excitatory neuron. b. Expression of selected marker genes used in cell type identification. The violin plot shows the distribution of normalized expression in nuclei in each cluster. Scale: normalized read counts. c. Heatmap plot shows the expression of the top 10 feature genes identified in each cluster. d. Dot plot shows the expression of astrocyte gene modules in each major cell type. e. Dot plot of the enrichR combined score for the top enriched GO terms for astrocyte gene modules M12 and M14. f. WGCNA dendrogram of Cortical Ex gene modules. g. Dot plot shows the expression of Cortical Ex gene modules in each major cell type. h. UMAP plots of module hub gene expression score for Cortical Ex gene modules M1–4. i. Co-expression plots of the top 25 module genes for Cortical Ex gene modules M1–4. j. Dot plot of the enrichR combined score for the top enriched GO terms for cortical excitatory neuron gene modules M1–4.
Extended Data Fig. 3 Maturation of astrocytes in glia-enriched cortical organoids.
a. Immunostaining of five-month-old glia-enriched cortical organoids: GFAP::GFP AAV-labeled astrocytes (GFP, green), presynaptic vesicles (SV2, gray), postsynaptic density (PSD95, magenta). Inset, an enlarged view of astrocyte process and synapses. Scale bars, 20 μm and 2 μm (inset). b. Immunostaining of five-month-old glia-enriched cortical organoids: GFAP::GFP AAV-labeled astrocytes (GFP, green), glutamate transporter (EAAT2, magenta). Scale bar, 20 μm. c. Immunostaining of five-month-old glia-enriched cortical organoids: GFAP::GFP AAV-labeled astrocytes (GFP, green) and matricellular protein (HEVIN, magenta). Scale bar, 5 μm. d. Immunostaining of five-month-old glia-enriched cortical organoids: GFAP::GFP AAV-labeled astrocytes (GFP, green), inward-rectifier potassium channel (Kir4.1, magenta), postsynaptic density protein (PSD95, cyan). Scale bar, 2 μm. e. Immunostaining of five-month-old glia-enriched cortical organoids: astrocytes (hGFAP, green), Connexin 43 (CX43, magenta). Inset, an enlarged view of astrocyte processes and expression of Connexin 43. Scale bars, 20 μm and 2 μm (inset). f. Quantification of glutamate uptake of neural progenitor cells (NPCs, n = 8) and astrocytes purified from five-month-old glia-enriched cortical organoids derived from Hues6 (n = 8), iPSC822 (n = 8) and H9 (n = 7) stem cell lines. Each dot represents one independent experiment. Bars, mean ± s.e.m.
Extended Data Fig. 4 Formation of anatomically defined morphological subclasses of human astrocytes in engrafted glia-enriched cortical organoids.
a. Immunostaining of glia-enriched cortical organoid transplants: human nuclear antigen (HuNu, Cyan), astrocytes (GFAP, green), neurons (NeuN, magenta). Scale bar, 20 μm. b. Quantification of the percentage of NeuN+ HuNu+ cells among NeuN+ cells (left) or huGFAP+ GFAP+ cells among GFAP+ cells (right) in the transplants. n = 4 transplants. Bars, mean ± s.e.m. c. Immunostaining of the center (left) and border (right) of glia-enriched cortical organoid transplants: human astrocytes (hGFAP, green), astrocytes (GFAP, magenta). Scale bars, 50 μm. d. Immunostaining of the center (left) and border (right) of glia-enriched cortical organoid transplants: oligodendrocyte progenitor cells (PDGFRα, green), human nuclear antigen (HuNu, magenta). Inset, enlarged views of human oligodendrocyte progenitor cells. Scale bars, 50 μm and 10 μm (inset). e. Immunostaining of human interlaminar, protoplasmic and fibrous astrocytes in glia-enriched cortical organoid transplants: human astrocytes (hGFAP, green), white matter (myelin basic protein or MBP, gray). Scale bars, 20 μm. f. Immunostaining of glia-enriched cortical organoid transplants: human cells (GFP, green), astrocytes (S100B, gray; CD44, magenta). Scale bar, 100 μm. g. Immunostaining of the top, center and bottom regions of the transplants: astrocytes (CD44, magenta), white matter (myelin basic protein or MBP, gray). Scale bars, 20 μm.
Extended Data Fig. 5 Spatial transcriptome profiling of layer-specific subclasses of human astrocytes in engrafted glia-enriched cortical organoids.
a. Immunofluorescent staining images on the left showing hGFAP in yellow, SOX9 in magenta and CYTO-13 in blue, along with the cell segmentation of SOX9+/CYTO-13+ cells depicted in magenta within a selected ROI on the right. Scale bars, 100 μm. b. Box plots depicting the normalized expression levels of selected genes in each group (pial = 10 ROIs, cortex = 16 ROIs, WM = 12 ROIs). Centerline, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers. c. Bubble plots depicting top significant terms identified from GSEA using a weighted Kolmogorov–Smirnov test. NES, normalized enrichment score. Significant thresholds set at adjusted p value < 0.1.
Extended Data Fig. 6 Close association of human protoplasmic astrocytes with synapses and host vasculature in engrafted glia-enriched cortical organoids.
a. Immunostaining of transplants: human astrocytes (hGFAP, green), glutamate transporters (EAAT2, magenta). Scale bar, 20 μm. b. Immunostaining of transplants: human astrocytes (hGFAP, green), matricellular protein (HEVIN, gray), postsynaptic density (PSD95, magenta). Scale bar, 20 μm. c. Electron micrograph of a multi-synaptic bouton in an eight-month-old transplant. Arrowheads: synapses. Scale bar, 1 μm. d. 3D reconstruction of serial section electron microscope images of synaptic structures in an eight-month-old transplant. Presynaptic bouton (orange), postsynaptic density (magenta), dendritic spine (blue). Scale bar, 1 μm. e. Electron micrograph of a synapse within the transplant. Left, presynaptic bouton (orange), postsynaptic density (PSD, magenta), dendritic spine (blue) and astrocytic processes (AP, green). Right, original image. Scale bar, 1 μm. f. Immunostaining of transplants at 2 (left panel) or 7 months (right panel) post-transplantation: blood vessels (Ly6C, cyan). Scale bars, 100 μm. g. Ly6C+ area (left) or vessel diameter (right) in transplants at 2 months (n = 4 transplants) or 7 months (n = 4 transplants) post-transplantation versus contralateral mouse cortex (Ms brain; n = 8 mice). Bars, mean ± s.e.m. Two-sided t-test, **p = 0.001, ****p < 0.0001. h. Immunostaining of transplants: human nuclear antigen (HuNu, green), pericytes (NG2, magenta, indicated by arrowhead in the left image), monocytes (Ly6C, gray, indicated by arrowhead in the middle image), endothelial cells (CD31, magenta). Scale bars, 10 μm. i. Immunostaining of transplants (left) or host brains (right): human nuclear antigen (HuNu, green), microglia (IBA1, magenta). Inset: microglia. Scale bars, 50 μm and 10 μm (insets). j. Immunostaining of transplants: human astrocytes (hGFAP, green), inward-rectifier potassium channel (Kir4.1, magenta), blood vessels (Ly6C, cyan). Scale bar, 20 μm. k. Immunostaining of transplants: human astrocytes (hGFAP, green), glucose transporter (Glut1, magenta) and blood vessels (Ly6C, cyan). Scale bar, 20 μm. l. Confocal images of the transplant used in EM studies: blood vessels (DiI, magenta), transplant (GFP, green). Scale bar, 1 mm. m. 3D reconstruction of series SEM images: tight junction (yellow) and basement membrane (blue). Scale bar, 1 μm.
Extended Data Fig. 7 Transcriptomic analysis of integrated snRNA-seq datasets.
a. Heatmap plot shows the expression levels of the top 10 feature genes identified in each cluster. b. Dot plot shows the average expression of selected marker genes in nuclei in each cluster. c. Heatmap depicting the pairwise transcriptional cluster correlation of cell clusters using top 50 marker genes from each cluster in integrated organoid datasets with cell clusters in snRNA-seq datasets of multiple human cortical areas from Allen Brain Map. Immature, immature excitatory neuron; UL, upper layer cortical excitatory neuron; DL, deep-layer cortical excitatory neuron; In, inhibitory neuron; APC, astrocyte progenitor cell; Ast, astrocyte; OPC, oligodendrocyte progenitor cell; unknown, undetermined cell. d. Ridge plot shows the distributions of predicted age for nuclei derived from snRNA-seq data obtained from prefrontal cortex (PFC) samples ranging from 22 weeks of gestation (ga) to 40 years of age (ref. 34). e. UMAP plot of snRNA-seq data from PFC samples (ref. 34) colored by major cell types. f. UMAP plot of snRNA-seq data from PFC samples (ref. 34) with each nucleus colored by its donor age. g. UMAP plots of snRNA-seq data from 5-month-old organoids (5m_Org), 5-month-old transplants (5m_T), 6-month-old transplants (6m_T) and 8-month-old transplants (8m_T), projected on to the reference dataset, with each nucleus colored by the predicted age. h. Ridge plot shows the distributions of predicted age for UL nuclei from 5m_Org, 5m_T, 6m_T and 8m_T. Wilcoxon test (two-sided, ****, p < 0.0001; reference group, 5m_Org).
Extended Data Fig. 8 WGCNA and pseudotime analyses of integrated snRNA-seq datasets.
a. WGCNA dendrogram of gene modules constructed from astrocytes across different time points. b. Dot plot shows the expression levels of astrocyte gene modules across major cell type. c. WGCNA dendrogram of gene modules constructed from immature and UL excitatory neurons across different time points. d. Dot plot shows the expression levels of neuron gene modules in each major cell type. e. Co-expression plot of the top 25 hub genes (left) and UMAP plot (right) of module hub gene expression score for neuron gene modules M1. f. Violin plot of harmonized module score of neuron gene module M1 in UL excitatory neurons across different time points. Centerline, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers. Wilcoxon test (two-sided, ****, p < 0.0001; reference group, 5m_Org). g. UMAP dimensionality reduction displays the pseudotime trajectories of neurons from the integrated snRNA-seq. Each cell is colored by its pseudotime trajectory assignment. Pseudotime analysis separated by time point. One-sided Kolmogorov–Smirnov test (****, p < 2.2e-16; reference group, 5m_Org).
Extended Data Fig. 9 Transcriptome profiling reveals rapid activation of pro-inflammatory pathways in a subpopulation of astrocytes in vivo.
a. Flow cytometry sorting strategy for glial cells. b. UMAP plot (left) and violin plot (right) shows the expression of selected cell type marker genes. c. UMAP plots show the expression of selected genes. d. Violin plot shows the expression of selected genes in astrocyte clusters 1–4. Ast, astrocyte. e. Bar plot of the enrichR combined score for gene ontology terms of the top 100 significantly upregulated (top) and downregulated (bottom) genes following TNFα treatment in cluster 2 astrocytes. f. GSEA comparing TNFα-treated and saline-treated cluster 2 astrocytes. GO terms are shown. FDR, false discovery rate; NES, normalized enrichment score.
Extended Data Fig. 10 CD38 mediates inflammation-induced metabolic and mitochondrial stresses in human astrocytes.
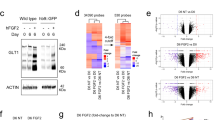
a. Immunostaining of in vitro glia-enriched cortical organoids at day 0, day 1 and day 2 post-TNFα treatment: astrocytes (hCD38, green; CD44 cyan), chemokines (CXCL10, magenta). Scale bars, 50 μm. b. Quantitative PCR analyses of selected genes in organoids (CTRL = 4; TNFα1d = 4; TNFα2d = 6). Each dot represents one organoid. Bars, mean ± s.d. c. Flow cytometry sorting strategy for GFAP::tdTomato+ cells. d. Quantitative PCR analyses of selected genes in sorted astrocytes (CTRL = 3 independent experiments; TNFα1d = 3 independent experiments). Each dot represents one independent experiment. Bars, mean ± s.d. Two-sided t-test, *p = 0.013555, ***p = 0.000948.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Video 1
Three-dimensional EM reconstruction of astrocytic endfeet structure.
Supplementary Tables
Supplementary Table 1: 10-week-old glia-enriched cortical organoid cell type cluster marker genes. Supplementary Table 2: Co-expression gene modules constructed using astrocytes in 10-week-old glia-enriched cortical organoids. Supplementary Table 3: Co-expression gene modules constructed using cortical excitatory neurons in 10-week-old glia-enriched cortical organoids. Supplementary Table 4: Differential gene expression across layer-specific astrocyte subclasses identified by GeoMx DSP. Supplementary Table 5: Differential gene expression across layer-specific astrocyte subclasses identified by GeoMx DSP. Supplementary Table 6: Differential gene expression across layer-specific astrocyte subclasses identified by GeoMx DSP. Supplementary Table 7: Integrated glia-enriched cortical organoid and transplant cell type cluster marker genes. Supplementary Table 8: Co-expression gene modules constructed using astrocytes in integrated glia-enriched cortical organoids and transplants. Supplementary Table 9: Co-expression gene modules constructed using immature and mature UL cortical excitatory neurons in integrated glia-enriched cortical organoids and transplants. Supplementary Table 10: Cell cluster marker genes from 8-month-old transplants treated with saline or TNFα. Supplementary Table 11: Primers used for quantitative PCR assay.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, M., Zhang, L., Novak, S.W. et al. Morphological diversification and functional maturation of human astrocytes in glia-enriched cortical organoid transplanted in mouse brain. Nat Biotechnol (2024). https://doi.org/10.1038/s41587-024-02157-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41587-024-02157-8