Abstract
Purpose
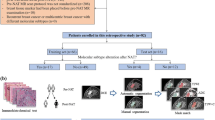
To improve the workflow of total marrow and lymphoid irradiation (TMLI) by enhancing the delineation of organs at risk (OARs) and clinical target volume (CTV) using deep learning (DL) and atlas-based (AB) segmentation models.
Materials and methods
Ninety-five TMLI plans optimized in our institute were analyzed. Two commercial DL software were tested for segmenting 18 OARs. An AB model for lymph node CTV (CTV_LN) delineation was built using 20 TMLI patients. The AB model was evaluated on 20 independent patients, and a semiautomatic approach was tested by correcting the automatic contours. The generated OARs and CTV_LN contours were compared to manual contours in terms of topological agreement, dose statistics, and time workload. A clinical decision tree was developed to define a specific contouring strategy for each OAR.
Results
The two DL models achieved a median [interquartile range] dice similarity coefficient (DSC) of 0.84 [0.71;0.93] and 0.85 [0.70;0.93] across the OARs. The absolute median Dmean difference between manual and the two DL models was 2.0 [0.7;6.6]% and 2.4 [0.9;7.1]%. The AB model achieved a median DSC of 0.70 [0.66;0.74] for CTV_LN delineation, increasing to 0.94 [0.94;0.95] after manual revision, with minimal Dmean differences. Since September 2022, our institution has implemented DL and AB models for all TMLI patients, reducing from 5 to 2 h the time required to complete the entire segmentation process.
Conclusion
DL models can streamline the TMLI contouring process of OARs. Manual revision is still necessary for lymph node delineation using AB models.

Similar content being viewed by others
References:
Paix A, Antoni D, Waissi W, Ledoux M-P, Bilger K, Fornecker L et al (2018) Total body irradiation in allogeneic bone marrow transplantation conditioning regimens: a review. Crit Rev Oncol Hematol 123:138–148. https://doi.org/10.1016/j.critrevonc.2018.01.011
Wong JYC, Filippi AR, Scorsetti M, Hui S, Muren LP, Mancosu P (2020) Total marrow and total lymphoid irradiation in bone marrow transplantation for acute leukaemia. Lancet Oncol 21:e477–e487. https://doi.org/10.1016/S1470-2045(20)30342-9
Wilkie JR, Tiryaki H, Smith BD, Roeske JC, Radosevich JA, Aydogan B (2008) Feasibility study for linac-based intensity modulated total marrow irradiation: linac-based intensity modulated total marrow irradiation. Med Phys 35:5609–5618. https://doi.org/10.1118/1.2990779
Yeginer M, Roeske JC, Radosevich JA, Aydogan B (2011) Linear accelerator-based intensity-modulated total marrow irradiation technique for treatment of hematologic malignancies: a dosimetric feasibility study. Int J Radiat Oncol Biol Phys 79:1256–1265. https://doi.org/10.1016/j.ijrobp.2010.06.029
Fogliata A, Cozzi L, Clivio A, Ibatici A, Mancosu P, Navarria P et al (2011) Preclinical assessment of volumetric modulated arc therapy for total marrow irradiation. Int J Radiat Oncol Biol Phys 80:628–636. https://doi.org/10.1016/j.ijrobp.2010.11.028
Han C, Schultheisss TE, Wong JYC (2012) Dosimetric study of volumetric modulated arc therapy fields for total marrow irradiation. Radiother Oncol 102:315–320. https://doi.org/10.1016/j.radonc.2011.06.005
Aydogan B, Yeginer M, Kavak GO, Fan J, Radosevich JA, Gwe-Ya K (2011) Total marrow irradiation with rapidarc volumetric arc therapy. Int J Radiat Oncol Biol Phys 81:592–599. https://doi.org/10.1016/j.ijrobp.2010.11.035
Dei D, Lambri N, Stefanini S, Vernier V, Brioso RC, Crespi L et al (2023) Internal guidelines for reducing lymph node contour variability in total marrow and lymph node irradiation. Cancers 15:1536. https://doi.org/10.3390/cancers15051536
Cha E, Elguindi S, Onochie I, Gorovets D, Deasy JO, Zelefsky M et al (2021) Clinical implementation of deep learning contour autosegmentation for prostate radiotherapy. Radiother Oncol 159:1–7. https://doi.org/10.1016/j.radonc.2021.02.040
van der Veen J, Willems S, Bollen H, Maes F, Nuyts S (2020) Deep learning for elective neck delineation: More consistent and time efficient. Radiother Oncol 153:180–188. https://doi.org/10.1016/j.radonc.2020.10.007
Ma C, Zhou J, Xu X, Guo J, Han M, Gao Y et al (2022) Deep learning-based auto-segmentation of clinical target volumes for radiotherapy treatment of cervical cancer. J Appl Clin Med Phys. https://doi.org/10.1002/acm2.13470
Byun HK, Chang JS, Choi MS, Chun J, Jung J, Jeong C et al (2021) Evaluation of deep learning-based autosegmentation in breast cancer radiotherapy. Radiat Oncol 16:203. https://doi.org/10.1186/s13014-021-01923-1
Kim H, Jung J, Kim J, Cho B, Kwak J, Jang JY et al (2020) Abdominal multi-organ auto-segmentation using 3D-patch-based deep convolutional neural network. Sci Rep 10:6204. https://doi.org/10.1038/s41598-020-63285-0
Amjad A, Xu J, Thill D, Lawton C, Hall W, Awan MJ et al (2022) General and custom deep learning autosegmentation models for organs in head and neck, abdomen, and male pelvis. Med Phys 49:1686–1700. https://doi.org/10.1002/mp.15507
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444. https://doi.org/10.1038/nature14539
Zabel WJ, Conway JL, Gladwish A, Skliarenko J, Didiodato G, Goorts-Matthews L et al (2021) Clinical evaluation of deep learning and atlas-based auto-contouring of bladder and rectum for prostate radiation therapy. Pract Radiat Oncol 11:e80–e89. https://doi.org/10.1016/j.prro.2020.05.013
Wong WKH, Leung LHT, Kwong DLW (2016) Evaluation and optimization of the parameters used in multiple-atlas-based segmentation of prostate cancers in radiation therapy. BJR 89:20140732. https://doi.org/10.1259/bjr.20140732
Sharp G, Fritscher KD, Pekar V, Peroni M, Shusharina N, Veeraraghavan H et al (2014) Vision 20/20: perspectives on automated image segmentation for radiotherapy: perspectives on automated image segmentation for radiotherapy. Med Phys 41:050902. https://doi.org/10.1118/1.4871620
D’Aviero A, Re A, Catucci F, Piccari D, Votta C, Piro D et al (2022) Clinical validation of a deep-learning segmentation software in head and neck: an early analysis in a developing radiation oncology center. IJERPH 19:9057. https://doi.org/10.3390/ijerph19159057
Radici L, Ferrario S, Borca VC, Cante D, Paolini M, Piva C et al (2022) Implementation of a commercial deep learning-based auto segmentation software in radiotherapy: evaluation of effectiveness and impact on workflow. Life 12:2088. https://doi.org/10.3390/life12122088
Chen W, Wang C, Zhan W, Jia Y, Ruan F, Qiu L et al (2021) A comparative study of auto-contouring softwares in delineation of organs at risk in lung cancer and rectal cancer. Sci Rep 11:23002. https://doi.org/10.1038/s41598-021-02330-y
Duan J, Bernard M, Downes L, Willows B, Feng X, Mourad WF et al (2022) Evaluating the clinical acceptability of deep learning contours of prostate and organs-at-risk in an automated prostate treatment planning process. Med Phys 49:2570–2581. https://doi.org/10.1002/mp.15525
Chen X, Sun S, Bai N, Han K, Liu Q, Yao S et al (2021) A deep learning-based auto-segmentation system for organs-at-risk on whole-body computed tomography images for radiation therapy. Radiother Oncol 160:175–184. https://doi.org/10.1016/j.radonc.2021.04.019
Shi J, Wang Z, Kan H, Zhao M, Xue X, Yan B (2022) Automatic segmentation of target structures for total marrow and lymphoid irradiation in bone marrow transplantation. In: 44th annual international conference of the IEEE engineering in medicine & biology society (EMBC), Glasgow, Scotland, United Kingdom: IEEE; 2022, p. 5025–9. https://doi.org/10.1109/EMBC48229.2022.9871824
Mancosu P, Navarria P, Muren LP, Castagna L, Reggiori G, Clerici E et al (2021) Development of an immobilization device for total marrow irradiation. Pract Radiat Oncol 11:e98-105. https://doi.org/10.1016/j.prro.2020.02.012
Mancosu P, Navarria P, Castagna L, Roggio A, Pellegrini C, Reggiori G et al (2012) Anatomy driven optimization strategy for total marrow irradiation with a volumetric modulated arc therapy technique. J Appl Clin Med Phys 13:138–147. https://doi.org/10.1120/jacmp.v13i1.3653
Mancosu P, Navarria P, Castagna L, Reggiori G, Sarina B, Tomatis S et al (2013) Interplay effects between dose distribution quality and positioning accuracy in total marrow irradiation with volumetric modulated arc therapy: TMI by VMAT: positioning uncertainties. Med Phys 40:111713. https://doi.org/10.1118/1.4823767
Mancosu P, Navarria P, Castagna L, Reggiori G, Stravato A, Gaudino A et al (2015) Plan robustness in field junction region from arcs with different patient orientation in total marrow irradiation with VMAT. Physica Med 31:677–682. https://doi.org/10.1016/j.ejmp.2015.05.012
Lambri N, Dei D, Hernandez V, Castiglioni I, Clerici E, Crespi L et al (2022) Automatic planning of the lower extremities for total marrow irradiation using volumetric modulated arc therapy. Strahlenther Onkol. https://doi.org/10.1007/s00066-022-02014-0
Sarina B, Mancosu P, Navarria P, Bramanti S, Mariotti J, De Philippis C et al (2021) Nonmyeloablative conditioning regimen including low-dose total marrow/lymphoid irradiation before haploidentical transplantation with post-transplantation cyclophosphamide in patients with advanced lymphoproliferative diseases. Transplant Cell Ther 27:492.e1-492.e6. https://doi.org/10.1016/j.jtct.2021.03.013
Jingnan Jia. Jingnan-Jia/segmentation_metrics: v1.1.3. Zenodo; 2022. https://doi.org/10.5281/ZENODO.7264630.
Panchal A, Pyup.Io Bot, Couture G, Gertsikkema, Galler N, Hideki_Nakamoto, (2019) dicompyler/dicompyler-core v0.5.5. Zenodo; 2019. https://doi.org/10.5281/ZENODO.3236628
Robert C, Munoz A, Moreau D, Mazurier J, Sidorski G, Gasnier A et al (2021) Clinical implementation of deep-learning based auto-contouring tools–Experience of three French radiotherapy centers. Cancer/Radiothérapie 25:607–616. https://doi.org/10.1016/j.canrad.2021.06.023
Chen M, Wu S, Zhao W, Zhou Y, Zhou Y, Wang G (2022) Application of deep learning to auto-delineation of target volumes and organs at risk in radiotherapy. Cancer/Radiothérapie 26:494–501. https://doi.org/10.1016/j.canrad.2021.08.020
Francolini G, Desideri I, Stocchi G, Salvestrini V, Ciccone LP, Garlatti P et al (2020) Artificial Intelligence in radiotherapy: state of the art and future directions. Med Oncol 37:50. https://doi.org/10.1007/s12032-020-01374-w
Vinod SK, Min M, Jameson MG, Holloway LC (2016) A review of interventions to reduce inter-observer variability in volume delineation in radiation oncology. J Med Imaging Radiat Oncol 60:393–406. https://doi.org/10.1111/1754-9485.12462
Casati M, Piffer S, Calusi S, Marrazzo L, Simontacchi G, Di Cataldo V et al (2022) Clinical validation of an automatic atlas-based segmentation tool for male pelvis CT images. J Applied Clin Med Phys. https://doi.org/10.1002/acm2.13507
Wong J, Fong A, McVicar N, Smith S, Giambattista J, Wells D et al (2020) Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother Oncol 144:152–158. https://doi.org/10.1016/j.radonc.2019.10.019
Watkins WT, Qing K, Han C, Hui S, Liu A (2022) Auto-segmentation for total marrow irradiation. Front Oncol 12:970425. https://doi.org/10.3389/fonc.2022.970425
Vandewinckele L, Claessens M, Dinkla A, Brouwer C, Crijns W, Verellen D et al (2020) Overview of artificial intelligence-based applications in radiotherapy: recommendations for implementation and quality assurance. Radiother Oncol 153:55–66. https://doi.org/10.1016/j.radonc.2020.09.008
Brouwer CL, Steenbakkers RJHM, Bourhis J, Budach W, Grau C, Grégoire V et al (2015) CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines. Radiother Oncol 117:83–90. https://doi.org/10.1016/j.radonc.2015.07.041
Funding
This work was funded by the Italian Ministry of Health, Grant AuToMI (GR-2019-12370739).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by DD, NL, LC, RCB, DL, and PM The first draft of the manuscript was written by DD and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no conflict of interests to disclose.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Ethics Committee of IRCCS Humanitas Research Hospital (ID 2928, 26 January 2021). ClinicalTrials.gov identifier: NCT04976205.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dei, D., Lambri, N., Crespi, L. et al. Deep learning and atlas-based models to streamline the segmentation workflow of total marrow and lymphoid irradiation. Radiol med 129, 515–523 (2024). https://doi.org/10.1007/s11547-024-01760-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11547-024-01760-8