Abstract
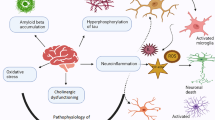
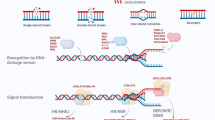
Genomic instability is a key driving force for the development and progression of many age-related neurodegenerative diseases and central nervous system (CNS) cancers. Recently, the cytosolic DNA sensor, cyclic GMP-AMP synthase (cGAS), has been shown to detect and respond to self-DNA accumulation resulting from DNA damaging insults in peripheral cell types. cGAS has been shown to be important in the responses of microglia to DNA viruses and amyloid beta, and we have reported that it underlies the responses of human microglia to exogenous DNA. However, the role of this cytosolic sensor in the detection of self-DNA by glia is poorly understood and its ability to mediate the cellular responses of human microglia to genotoxic DNA damage has not been established. Here, we describe the ability of ionizing radiation and oxidative stress to elicit genomic DNA damage in human microglial cells and to stimulate the production of key inflammatory mediators by these cells in an NF-kB dependent manner. Importantly, we have utilized CRISPR/Cas9 and siRNA-mediated knockdown approaches and a pharmacological inhibitor of the cGAS adaptor protein stimulator of interferon genes (STING) to demonstrate that the cGAS-STING pathway plays a critical role in the generation of these microglial immune responses to such genotoxic insults. Together, these studies support the notion that cGAS mediates the detection of cytosolic self-DNA by microglia, providing a potential mechanism linking genomic instability to the development of CNS cancers and neurodegenerative disorders.






Similar content being viewed by others
Availability of Data and Material
The data used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
Zhu, L.-S., D.-Q. Wang, K. Cui, D. Liu, and L.-Q. Zhu. 2019. Emerging perspectives on DNA double-strand breaks in neurodegenerative diseases. Current Neuropharmacology 17: 1146–1157.
Coppedè, F., and L. Migliore. 2015. DNA damage in neurodegenerative diseases. Mutation Research - Fundamental and Molecular Mechanisms of Mutagenesis 776: 84–97.
Horn, V., and A. Triantafyllopoulou. 2018. DNA damage signaling and polyploid macrophages in chronic inflammation. Current Opinion in Immunology 50: 55–63.
Taffoni, C., A. Steer, J. Marines, H. Chamma, I.K. Vila, and N. Laguette. 2021. nucleic acid immunity and DNA damage response: new friends and old foes. Frontiers in Immunology 12: 1–10.
Li, T., and Z.J. Chen. 2018. The cGAS-cGAMP-STI NG pathway connects DNA damage to inflammation, senescence, and cancer. Journal of Experimental Medicine 215: 1287–1299.
Krupina, K., A. Goginashvili, and D.W. Cleveland. 2021. Causes and consequences of micronuclei. Current Opinion in Cell Biology 70: 91–99.
Miller, K.N., S.G. Victorelli, H. Salmonowicz, N. Dasgupta, T. Liu, J.F. Passos, et al. 2021. Cytoplasmic DNA: sources, sensing, and role in aging and disease. Cell 184: 5506–5526.
Sun, L., J. Wu, F. Du, X. Chen, and Z.J. Chen. 1979. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013 (339): 786–791.
Ablasser, A., M. Goldeck, T. Cavlar, T. Deimling, G. Witte, I. Röhl, et al. 2013. CGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498: 380–384.
Sun, W., Y. Li, L. Chen, H. Chen, F. You, X. Zhou, et al. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proceedings of the National Academy of Sciences 106: 8653–8658.
Wu, J., L. Sun, X. Chen, F. Du, H. Shi, C. Chen, et al. 1979. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013 (339): 826–830.
Zhong, B., Y. Yang, S. Li, Y.Y. Wang, Y. Li, F. Diao, et al. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29: 538–550.
De Gaetano, A., K. Solodka, G. Zanini, V. Selleri, A.V. Mattioli, M. Nasi, et al. 2021. Molecular mechanisms of mtDNA-mediated inflammation. Cells 10: 2898.
Gao, D., T. Li, X.-D. Li, X. Chen, Q.-Z. Li, M. Wight-Carter, et al. 2015. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proceedings of the National Academy of Sciences 112: E5699–E5705.
Mackenzie, K.J., P. Carroll, C.-A.A. Martin, O. Murina, A. Fluteau, D.J. Simpson, et al. 2017. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548: 461–465.
Kumar, V. 2021. The trinity of cGAS, TLR9, and ALRs guardians of the cellular galaxy against host-derived self-DNA. Frontiers in Immunology 11: 1–31.
Jakobs, C., S. Perner, and V. Hornung. 2015. AIM2 drives joint inflammation in a self-DNA triggered model of chronic polyarthritis. PLoS One 10: e0131702.
Cox, D.J., R.H. Field, D.G. Williams, M. Baran, A.G. Bowie, C. Cunningham, et al. 2015. DNA sensors are expressed in astrocytes and microglia in vitro and are upregulated during gliosis in neurodegenerative disease. Glia 63: 812–825.
Jeffries, A.M., and I. Marriott. 2017. Human microglia and astrocytes express cGAS-STING viral sensing components. Neuroscience Letters 658: 53–56.
Song, X., F. Ma, and K. Herrup. 2019. Accumulation of cytoplasmic DNA due to ATM deficiency activates the microglial viral response system with neurotoxic consequences. The Journal of Neuroscience 39: 6378–6394.
Jin, M., H. Shiwaku, H. Tanaka, T. Obita, S. Ohuchi, Y. Yoshioka, et al. 2021. Tau activates microglia via the PQBP1-cGAS-STING pathway to promote brain inflammation. Nature Communications 12: 6565.
Wu, P.J., Y.F. Hung, H.Y. Liu, and Y.P. Hsueh. 2017. Deletion of the inflammasome sensor Aim2 mitigates Aβ deposition and microglial activation but increases inflammatory cytokine expression in an Alzheimer disease mouse model. NeuroImmunoModulation 24: 29–39.
Rui, W.J., S. Li, L. Yang, Y. Liu, Y. Fan, Y.C. Hu, et al. 2022. Microglial AIM2 alleviates antiviral-related neuro-inflammation in mouse models of Parkinson’s disease. Glia 70: 2409–2425.
Ma, C., S. Li, Y. Hu, Y. Ma, Y. Wu, C. Wu, et al. 2021. AIM2 controls microglial inflammation to prevent experimental autoimmune encephalomyelitis. Journal of Experimental Medicine 218: e20201796.
Sterka, D., D.M. Rati, and I. Marriott. 2006. Functional expression of NOD2, a novel pattern recognition receptor for bacterial motifs, in primary murine astrocytes. Glia 53: 322–330.
Sterka, D., and I. Marriott. 2006. Characterization of nucleotide-binding oligomerization domain (NOD) protein expression in primary murine microglia. Journal of Neuroimmunology 179: 65–75.
Furr, S.R., V. Chauhan, D. Sterka, V. Grdzelishvili, and I. Marriott. 2008. Characterization of retinoic acid-inducible gene-I expression in primary murine glia following exposure to vesicular stomatitis virus. Journal of Neurovirology 14: 503–513.
Gao, P., M. Ascano, Y. Wu, W. Barchet, B.L. Gaffney, T. Zillinger, et al. 2013. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153: 1094–1107.
Orzalli, M.H., N.M. Broekema, B.A. Diner, D.C. Hancks, N.C. Elde, I.M. Cristea, et al. 2015. CGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proceedings of the National Academy of Sciences 112: E1773–E1781.
Ruangkiattikul, N., A. Nerlich, K. Abdissa, S. Lienenklaus, A. Suwandi, N. Janze, et al. 2017. cGAS-STING-TBK1-IRF3/7 induced interferon-β contributes to the clearing of non tuberculous mycobacterial infection in mice. Virulence 8: 1303–1315.
Aguirre, S., P. Luthra, M.T. Sanchez-Aparicio, A.M. Maestre, J. Patel, F. Lamothe, et al. 2017. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nature Microbiology 2: 1–11.
Wong, E.B., B. Montoya, M. Ferez, C. Stotesbury, and L.J. Sigal. 2019. Resistance to ectromelia virus infection requires cGAS in bone marrow-derived cells which can be bypassed with cGAMP therapy. PLoS Pathogens 15: e1008239.
Wu, Y., K. Song, W. Hao, J. Li, L. Wang, and S. Li. 2022. Nuclear soluble cGAS senses double-stranded DNA virus infection. Communications Biology 5: 1–13.
Reinert, L.S., K. Lopušná, H. Winther, C. Sun, M.K. Thomsen, R. Nandakumar, et al. 2016. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defence in the CNS. Nature Communications 7: 13348.
Zhang, Q., Z. Tang, R. An, L. Ye, and B. Zhong. 2020. USP29 maintains the stability of cGAS and promotes cellular antiviral responses and autoimmunity. Cell Research 30: 914–927.
Fruhwürth, S., L.S. Reinert, C. Öberg, M. Sakr, M. Henricsson, H. Zetterberg, et al. 2023. TREM2 is down-regulated by HSV1 in microglia and involved in antiviral defense in the brain. https://www.science.org.
Hou, Y., Y. Wei, S. Lautrup, B. Yang, Y. Wang, S. Cordonnier, et al. 2021. NAD+ supplementation reduces neuroinflammation and cell senescence in a transgenic mouse model of Alzheimer’s disease via cGAS–STING. Proceedings of the National Academy of Sciences 118: e2011226118. https://pnas.org/doi/full/10.1073/pnas.2011226118.
Xie, X., G. Ma, X. Li, J. Zhao, Z. Zhao, and J. Zeng. 2023. Activation of innate immune cGAS-STING pathway contributes to Alzheimer’s pathogenesis in 5×FAD mice. Nature Aging 3: 202–212. https://www.nature.com/articles/s43587-022-00337-2.
Suptela, A.J., and I. Marriott. 2023. Cytosolic DNA sensors and glial responses to endogenous DNA. Frontiers in Immunology 14: 1130172.
Gulen, M.F., N. Samson, A. Keller, M. Schwabenland, C. Liu, S. Glück, et al. 2023. cGAS–STING drives ageing-related inflammation and neurodegeneration. Nature 620: 374–380.
Johnson, M.B., J.R. Halman, A.R. Burmeister, S. Currin, E.F. Khisamutdinov, K.A. Afonin, et al. 2020. Retinoic acid inducible gene-I mediated detection of bacterial nucleic acids in human microglial cells. Journal of Neuroinflammation 17: 1–14.
Burmeister, A.R., M.B. Johnson, V.S. Chauhan, M.J. Moerdyk-Schauwecker, A.D. Young, I.D. Cooley, et al. 2017. Human microglia and astrocytes constitutively express the neurokinin-1 receptor and functionally respond to substance P. Journal of Neuroinflammation 14: 245.
Jeffries, A.M., Nitika, A.W. Truman, and I. Marriott. 2020. The intracellular DNA sensors cGAS and IFI16 do not mediate effective antiviral immune responses to HSV-1 in human microglial cells. Journal of Neurovirology 26: 544–555. https://link.springer.com/10.1007/s13365-020-00852-1.
Garcia-Mesa, Y., T.R. Jay, M.A. Checkley, B. Luttge, C. Dobrowolski, S. Valadkhan, et al. 2017. Immortalization of primary microglia: a new platform to study HIV regulation in the central nervous system. Journal of Neurovirology 23: 47–66.
Moravan, M.J., J.A. Olschowka, J.P. Williams, and M.K. O’Banion. 2011. Cranial irradiation leads to acute and persistent neuroinflammation with delayed increases in T-cell infiltration and CD11c expression in C57BL/6 mouse brain. Radiation Research 176: 459–473.
Mueller, S., G. Millonig, and G.N. Waite. 2009. The GOX/CAT system: a novel enzymatic method to independently control hydrogen peroxide and hypoxia in cell culture. Advances in Medical Sciences 54: 121–135.
Porciani, D., L. Tedeschi, L. Marchetti, L. Citti, V. Piazza, F. Beltram, et al. 2015. Aptamer-mediated codelivery of doxorubicin and NF-κB decoy enhances chemosensitivity of pancreatic tumor cells. Molecular Therapy-Nucleic Acids 4: e235.
Ke, W., E. Hong, R.F. Saito, M.C. Rangel, J. Wang, M. Viard, et al. 2019. RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-κB in human cells. Nucleic Acids Research 47: 1350–1361.
Shlyakhtenko, L.S., A.A. Gall, A. Filonov, Z. Cerovac, A. Lushnikov, and Y.L. Lyubchenko. 2003. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy 97: 279–287.
Ran, F.A., P.D. Hsu, J. Wright, V. Agarwala, D.A. Scott, and F. Zhang. 2013. Genome engineering using the CRISPR-Cas9 system. Nature Protocols 8: 2281–2308.
Einor, D., A. Bonisoli-Alquati, D. Costantini, T.A. Mousseau, and A.P. Møller. 2016. Ionizing radiation, antioxidant response and oxidative damage: a meta-analysis. Science of the Total Environment 548–549: 463–471.
Maekawa, H., T. Inoue, H. Ouchi, T.M. Jao, R. Inoue, H. Nishi, et al. 2019. Mitochondrial damage causes inflammation via cGAS-STING signaling in acute kidney injury. Cell Reports 29: 1261–1273.e6.
Liu, S., M. Feng, and W. Guan. 2016. Mitochondrial DNA sensing by STING signaling participates in inflammation, cancer and beyond. International Journal of Cancer 139: 736–741.
Wu, Z., A.G. Sainz, and G.S. Shadel. 2021. Mitochondrial DNA: cellular genotoxic stress sentinel. Trends in Biochemical Sciences 46: 812–821.
de Oliveira Mann, C.C., and P.J. Kranzusch. 2017. cGAS conducts micronuclei DNA surveillance. Trends in Cell Biology 27: 697–698.
Zhao, Y., B. Liu, L. Xu, S. Yu, J. Fu, J. Wang, et al. 2021. ROS-induced mtDNA release: the emerging messenger for communication between neurons and innate immune cells during neurodegenerative disorder progression. Antioxidants 10: 1917.
Motwani, M., and K.A. Fitzgerald. 2017. cGAS Micro-manages genotoxic stress. Immunity 47: 616–617.
Bakhoum, S.F., B. Ngo, A.M. Laughney, J.A. Cavallo, C.J. Murphy, P. Ly, et al. 2018. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553: 467–472.
Sharma, M., S. Rajendrarao, N. Shahani, U.N. Ramírez-Jarquín, and S. Subramaniam. 2020. Cyclic GMP-AMP synthase promotes the inflammatory and autophagy responses in Huntington disease. Proceedings of the National Academy of Sciences 117: 15989–15999.
Mohr, L., E. Toufektchan, P. von Morgen, K. Chu, A. Kapoor, and J. Maciejowski. 2021. ER-directed TREX1 limits cGAS activation at micronuclei. Molecular Cell 81: 724–738.e9.
Harding, S.M., J.L. Benci, J. Irianto, D.E. Discher, A.J. Minn, and R.A. Greenberg. 2017. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548: 466–470.
Zhao, M., F. Wang, J. Wu, Y. Cheng, Y. Cao, X. Wu, et al. 2021. CGAS is a micronucleophagy receptor for the clearance of micronuclei. Autophagy 17: 3976–3991.
West, A.P., W. Khoury-Hanold, M. Staron, M.C. Tal, C.M. Pineda, S.M. Lang, et al. 2015. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520: 553–557.
Guo, Y., R. Gu, D. Gan, F. Hu, G. Li, and G. Xu. 2020. Mitochondrial DNA drives noncanonical inflammation activation via cGAS–STING signaling pathway in retinal microvascular endothelial cells. Cell Communication and Signaling 18: 1–12.
Huang, L.S., Z. Hong, W. Wu, S. Xiong, M. Zhong, X. Gao, et al. 2020. mtDNA activates cGAS signaling and suppresses the YAP-mediated endothelial cell proliferation program to promote inflammatory injury. Immunity 52: 475–486.e5.
Nadalutti, C.A., S. Ayala-Peña, and J.H. Santos. 2022. Mitochondrial DNA damage as driver of cellular outcomes. American Journal of Physiology. Cell Physiology 322: C136–C150.
Zhang, W., G. Li, R. Luo, J. Lei, Y. Song, B. Wang, et al. 2022. Cytosolic escape of mitochondrial DNA triggers cGAS-STING-NLRP3 axis-dependent nucleus pulposus cell pyroptosis. Experimental & Molecular Medicine 54: 129–142.
Chen, H., H. Chen, J. Zhang, Y. Wang, A. Simoneau, H. Yang, et al. 2020. cGAS suppresses genomic instability as a decelerator of replication forks. Science Advances 6: eabb8941.
Liu, H., H. Zhang, X. Wu, D. Ma, J. Wu, L. Wang, et al. 2018. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563: 131–136.
Jiang, H., X. Xue, S. Panda, A. Kawale, R.M. Hooy, F. Liang, et al. 2019. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO Journal 38: 1–17.
Hinkle, J.T., J. Patel, N. Panicker, S.S. Karuppagounder, D. Biswas, B. Belingon, et al. 2022. STING mediates neurodegeneration and neuroinflammation in nigrostriatal α-synucleinopathy. Proceedings of the National Academy of Sciences 119: 1–8.
Funding
This material is based upon work supported in whole or part by the North Carolina Biotechnology Center (to IM), the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R03 NS111260 (to CR), and by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35 GM139587 (to KAA).
Author information
Authors and Affiliations
Contributions
AJS carried out the in vitro experiments, specific capture ELISAs, immunoblot analyses, and performed data analysis. IM, CR, and SY conceived the study, contributed to the experimental design, and drafted the manuscript. KAA and YR designed, manufactured, and characterized the decoy DNA deploying constructs employed in these studies. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Suptela, A.J., Radwan, Y., Richardson, C. et al. cGAS Mediates the Inflammatory Responses of Human Microglial Cells to Genotoxic DNA Damage. Inflammation (2023). https://doi.org/10.1007/s10753-023-01946-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10753-023-01946-8