Abstract
Understanding how dietary polysaccharides affect mucin O-glycosylation and gut microbiota could provide various nutrition-based treatments. Here, the O-glycan profile of the colonic mucosa and gut microbiome were investigated in C57BL/6J mice fed six structurally diverse dietary polysaccharides and a mixture of six fibers. Dietary polysaccharides increased total O-glycans, mainly by stimulating neutral glycans. Highly branched arabinogalactan promoted terminally fucosylated core 1 O-glycans; whereas linear polysaccharides, including pectin, konjac glucomannan, inulin, and the fiber mixture, favored terminally di-fucosylated O-glycans. The last three polysaccharides also lowered the level of sulfated O-glycans and sialylated mono-fucosylated O-glycans. Varied monosaccharide composition in mixed polysaccharides had a synergistic beneficial effect, boosting fucosylated neutral glycans, decreasing acidic glycans, and stimulating microbial richness and diversity. Dietary polysaccharides containing arabinose and sulfate groups enhanced the relative abundances of Akkermansia and Muribaculaceae, respectively. The present comparison reveals the relationship between dietary polysaccharide structure, mucin O-glycan composition, and intestinal microorganisms.
Similar content being viewed by others
Introduction
The intestinal tract fulfills a double role by enabling the digestion and absorption of nutrients, as well as by influencing the host immune system1,2,3. Any damage to the intestinal barrier is associated with the occurrence and progression of intestinal or parenteral diseases4. The mucus layer lining the gut protects against potential biological, physical, and chemical attacks, while also providing adhesion sites and nutrient/energy sources in the form of specific O-glycans for symbiotic bacteria1,2,4,5,6,7.
Mucin (Muc2) is the major structural and functional constituent of the colonic mucus layer. More than 80% of its mass is constituted by O-glycans8,9. Mucin O-glycosylation begins with the binding of N-acetylgalactosamine (GalNAc) to the OH group of serine or threonine. The resulting Tn antigen is then extended through the addition of N-acetylglucosamine (GlcNAc), and GalNAc10. Core 1 Galβ1,3GalNAcα1-Ser/Thr and core 2 Galβ1,3(GlcNAcβ1,6)GalNAcα1-Ser/Thr structures are the main components of mouse Muc211. The core structure of O-glycans is further extended through the addition of fucose (Fuc), N-acetylneuraminic acid (Neu5Ac), GalNAc, Gal, and sulfated group, forming an even more complex structure12.
The biological functions of mucin are closely related to its diverse O-glycan structure. O-glycans confer viscoelasticity and hydration capacity8. The latter imparts gel-like properties and prevents proteases from degrading the protein backbone1. At the same time, mucin O-glycans provide colonization sites for probiotics, which prevent pathogen infection through site competition13. Colonization of the mucus layer by Akkermansia stimulates the production of intestinal Muc2 and host defenses, thereby preventing the settling of other pathogenic bacteria14,15. Finally, mucin O-glycans could serve as nutrients for specific intestinal bacteria, such as Akkermansia muciniphila, Bifidobacterium bifidum, Bacteroides thetaiotaomicron, and Ruminococcus torques, with the resulting oligosaccharides or monosaccharides then cross-fed to other intestinal bacteria to maintain microbial homeostasis16.
Mucin O-glycans impact the gut microbiota, meanwhile, the gut microbes and microbial metabolite production change intestinal mucin O-glycans1. Faecalibacterium prausnitzii and B. thetaiotaomicron, regulate the levels of glycosylation-related genes (e.g., c1galt1 and b4galt4) in mucin-secreting goblet cells17. Da Silva et al. found that oral administration of the probiotic Lactobacillus farciminis significantly reduced the expression of intestinal sialylated O-glycans (m/z 1331 and 1605) in irritable bowel syndrome18. The expression of glycosyltransferases is impacted by changes in bacterial compositions, which in turn affects the degree of mucin O-glycosylation. When feces from healthy mice were transplanted into germ-free mice, Clostridia, Lactobacillus, Bacteroides, and Rikenellaceae were the dominant bacteria in the mouse colon 8 weeks after the transplant, coinciding with the downregulation of St6GalNac1 and upregulation of Fut2 and C1GalT1 O-glycosyltransferases. This resulted in a significant decrease in sulfated O-glycans (m/z 667), but an increase in fucosylated O-glycans (m/z 975) in colonic mucin19. In addition, the expression of intestinal mucin O-glycans is influenced by short-chain fatty acids (SCFAs) generated by symbiotic microorganisms. Acetate alters mucin O-glycosylation by adjusting the expression of β1,3-galactosyltransferase and sialyltransferase17. Butyrate binds to antigen-presenting cells and induces the division of T regulatory cells, which causes the release of interleukin-1020. The latter then stimulates Muc2-related genes and alters the pattern of intestinal mucin O-glycans21. Therefore, this relationship is bidirectional between the gut bacteria and O-glycan of mucin.
Dietary intake plays a critical role in determining the composition of gut bacteria22. Dietary polysaccharides, which cannot be digested or absorbed in the stomach and small intestine, are fermented and metabolized by the microbiota of the colon. However, dietary polysaccharides originate from a broad range of sources and present complex chemical structures, their influence on the composition of intestinal bacteria varies23. Pectin from apples (RG-I) was found to favor Faecalibaculum and Lactobacillus spp. in the murine gut24,25; supplemented with Arabinogalactan from Lycium barbarum polysaccharides (LBP-3) improved the related abundance of the Prevotellaceae, Rikenellaceae26. Hence, the intake of different structures on dietary polysaccharides affects the colonization of intestinal microorganisms23.
In addition, the intake of dietary polysaccharides alters the mucus layer, mucin, and its O-glycans. Supplementation with naturally extracted LBPs and raspberry pectin (PEC), commercially available konjac glucomannan (KGM), and inulin (INU) results in a significantly thicker mucus layer26,27,28,29. Muc2 secretion in the mouse ileum and colon is promoted by supplementation with natural extracts of Dendrobium huoshanense polysaccharides, commercially available carrageenan, chondroitin sulfate (CS), and sodium alginate30,31. Addition of NutriKane and Benefiber polysaccharides (Gratuk Technologies Pty. Ltd., Macquarie Park, NSW, Australia) is associated with increased levels of fucosylated O-glycans and decreased levels of sialylated O-glycans in colonic mucin of obese mice12. The effect of crude or single dietary polysaccharides on intestinal mucus layer thickness or mucin expression has been studied mainly in disease models. A systematic glycomics comparison in healthy mice could reveal the inherent effect of dietary polysaccharides on mucin O-glycans.
We hypothesized that different structural dietary polysaccharides might affect the profile of intestinal Muc2 O-glycans by influencing the composition of the intestinal community. To verify this hypothesis, the present study investigated the effect of supplementing structurally diverse dietary polysaccharides on the mucin O-glycan profile and gut microbial composition of healthy C57BL/6J mice. The mice were supplemented for 9 weeks with KGM, Laminaria japonica polysaccharides (BSP), CS, L. barbarum arabinogalactan (LBP-AG), PEC, INU, and a mixture of six polysaccharides (MIX). The relationship between dietary polysaccharide structure, mucin O-glycan profile, and gut microbiota was investigated. Our results can lead to the development of dietary polysaccharides for use as health-promoting supplements that stimulate the intestinal barrier function.
Results
Effect of structurally diverse dietary polysaccharides on mouse colon morphology
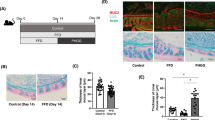
To explore how structurally diverse dietary polysaccharides affected colonic mucus O-glycans, C57BL/6J mice were allocated randomly to eight groups and fed either a normal diet (CON) or a normal diet supplemented with different polysaccharides (LBP-AG, CS, BSP, PEC, KGM, MIX, and INU) as shown in Fig. 1A. A detailed timeline of the experiment and the proportions of major nutrients in the diet are illustrated in Fig. 1B, C. Physiological parameters, such as body weight, spleen index, kidney index, and liver index (Fig. 1D) displayed no significant change on a weekly basis (P > 0.05), indicating that polysaccharide supplementation had no adverse effect on host health. Supplementation with dietary polysaccharides increased cecum weight more than in CON group, albeit without any significant difference across dietary polysaccharide groups (Fig. 1D).
A Structure of the six dietary polysaccharides used. B Timeline of the experiment. The mice were fed a normal diet or a diet containing the different polysaccharides (n = 10) for 18 weeks. The time and type of samples collected, and measurements performed are indicated. C Nutrient composition of each diet. Detailed nutritional information is provided in Supplementary Table 2. D Weekly changes in body weight per mouse (mean ± SD); organ index for the spleen, kidneys, and liver (mean ± SEM); and the weight of the caecum (mean ± SEM). Significance (P < 0.05) was determined using Duncan’s test (n = 10).
To evaluate whether polysaccharide administration altered mucus morphology, colon tissues were stained with hematoxylin and eosin, alcian blue, as well as periodic acid-schiff (PAS) in Fig. 2A–C. Mucin 2 of goblet cells was detected by immunofluorescence staining (Fig. 2D). The number of crypts and goblet cells in the mouse colon was calculated by gray-scale analysis of PAS images, and was significantly higher in dietary polysaccharide-supplemented animals compared to the control group (Fig. 2E, F), along with higher fluorescent immunostaining of Muc2 in the colon of mice (Fig. 2G). Again, there was no significant difference among dietary polysaccharide groups. To investigate the effect of dietary polysaccharides on gut barrier function in mice, tight junction proteins, including ZO-1, occludin, and claudin, were semi-quantified by western blotting (Supplementary Fig. 1A–D). All dietary polysaccharides caused a significant increase in the expression of these proteins in the colon (P < 0.05). The effect was particularly pronounced in the MIX group, indicating that its rich monosaccharide composition could improve the intestinal barrier of mice more than other dietary polysaccharides.
A Hematoxylin and eosin staining (scale bars, 200 μm). B PAS staining (scale bars, 200 μm). C Alcian blue staining (scale bars, 200 μm). D Fluorescent immunostaining of mucin 2 (scale bars, 20 μm). E Crypt number (mean ± SEM). F Number of goblet cells (mean ± SEM). G Quantification of mucin 2 in the goblet cells (mean ± SEM). Significance (P < 0.05) was determined using Duncan’s test (n = 6).
Supplementation with structurally diverse polysaccharides alters O-glycosylation of the colonic mucus layer
To determine the impact of structurally diverse polysaccharides on the profile of scraped colonic mucus layer O-glycans, the latter were released from equal amounts of intestinal mucin by the reductive β-elimination method. Equimolar amounts of β-cyclodextrin and 6’-sialyllactose were added as internal standards. Neutral and acidic O-glycans on Muc2 were analyzed in positive and negative ion mode, respectively. Relative quantification of colonic Muc2 O-glycans was performed using online tandem liquid chromatography (LC)-mass spectroscopy (MS). The structure of O-glycans was presumed based on MS (m/z) values, GlycoWorkbench version 2.1, and published reports3,12,32. Supplementary Table 3 lists total O-glycans, neutral and acidic O-glycans, non-fucosylated and fucosylated neutral O-glycans, sialylated and sulfated O-glycans, fucosylated O-glycans, both sulfated and sialylated O-glycans, as well as core 2, core 1, and core 4 glycans. Fifty-four types of O-glycans were identified in the CON group, with their LC-MS profile peaks reported in Supplementary Figs. 2 and 3, and the MS/MS profile of the isomer was shown in Supplementary Fig. 4. Even though the same O-glycans were identified in the CON and dietary polysaccharide groups, their relative levels differed significantly (P < 0.05).
Monosaccharide composition, presumed structure, m/z values, abundance peaks, and number of colonic Muc2 O-glycans in all groups are summarized in Supplementary Table 5. O-glycosylation levels were determined by comparative analysis, which revealed that intestinal mucin O-glycans increased upon supplementation with dietary polysaccharides (Fig. 3A). The total amount of O-glycans was significantly higher (P < 0.05) in the PEC group compared to other groups and 1.71 times higher than in the CON group. Further analysis demonstrated a significant increase in neutral O-glycans, but a decrease in acidic O-glycans upon supplementation with dietary polysaccharides (Fig. 3B, C). Again, neutral O-glycans were significantly more abundant in the PEC than in other groups (P < 0.05), and 2.97-fold more abundant than in the CON group. Acidic O-glycans were particularly scarce in the MIX group, whereby their level was 0.55 times lower than in CON group.
A–C Abundance of total (A), acidic (B), and neutral (C) O-glycans. (D–H) Subdivision according to specific modifications: non-fucosylated neutral O-glycans (D), fucosylated neutral O-glycans (E), sulfated O-glycans (F), sialylated O-glycans (G), and sulfated and sialylated acidic O-glycans (H). Data are presented as the mean ± SEM. Significance (P < 0.05) was determined using Duncan’s test (n = 5).
Neutral O-glycans were further assessed in terms of non-fucosylated and fucosylated glycans (Fig. 3D, E). Non-fucosylated O-glycans were more abundant upon supplementation with the different dietary polysaccharides, as shown in Fig. 4 for core 2 Gal-(GalNAc-6)GalNAcol (Number 3), whose content increased by ~2–4 fold. Compared to the CON group, the PEC group presented 14.73-fold higher levels of terminally non-fucosylated neutral O-glycans (H2N3, Number 9); whereas the MIX group led to 17.42-fold higher levels of core 2 O-glycans containing galactose (H2N2, Number 5). All dietary polysaccharide groups caused an increase in the content of terminally fucosylated neutral O-glycans, with the PEC and MIX groups achieving 2.24 times and 1.80 times higher levels, respectively, than the CON group (Fig. 3E). Fucosyltransferase 2 (Fut2), which mediates the addition of Fuc to Gal via an α-1-2 linkage, is the only fucose glycosyltransferase in mice3. The expression of Fut2 in the colon of mice was determined in different dietary polysaccharide groups (Supplementary Fig. 1A, E). Fut2 expression was similar to that of fucosylated O-glycans on mucin, with the PEC group again exhibiting significantly higher levels than other groups.
As shown in Fig. 4, the LBP group exhibited 13.13 times higher levels of the terminally mono-fucosylated core 1 O-glycan (Fucα1,2Galβ1,3GalNAcol, Number 2); PEC and MIX increased by 4.92 and 4.26 times the level of terminally di-fucosylated F2H2N2 (Number 11); and CS and PEC augmented by 1.19 and 2.51 times the amount of F1H1N2, another terminally fucosylated O-glycan (Number 2). Consistent with our results, supplementation with corn-resistant starch boosts the activity of fucosyltransferase in the small intestine of normal rats and, consequently, the amount of fucosylated O-glycans33.
Acidic O-glycans were further analyzed in terms of sulfated O-glycans, sialylated O-glycans, and both sulfated and sialylated O-glycans (Fig. 3F–H). All three O-glycan types were less abundant following dietary polysaccharide supplementation (P < 0.05). Furthermore, a decrease in O-glycans with a terminal sulfate group (S1H1N2, Number 25) and sulfated O-glycans with a terminal fucose (S1F1H1N2, Numbers 26 and 27) contributed to fewer sulfated O-glycans, as outlined in Figs. 3F–H and 4. Sulfated O-glycan content was much lower (46.5%) in the MIX group compared to all other groups (P < 0.05). Instead, sialylated O-glycans were particularly scarce (66.7%) in the KGM group, and included H2N3A1, H1N3A1, and another 10 glycoforms. Lower levels were detected also in the INU (52.7%) and MIX (47.3%) groups. The KGM and INU groups displayed a similar drop in sialylated O-glycans harboring a terminal fucose (F1H2N2A1, Numbers 39 and 40; F1H3N2A1, Numbers 44 and 45). For both sulfated and sialylated O-glycans, the INU and KGM groups presented comparable reductions of 85.2% and 83.4%, respectively, whereas the CS and MIX groups displayed 69.6% and 59.3% lower amounts, respectively. Overall, the MIX, INU, and KGM groups exhibited significantly (P < 0.05) lower levels of sulfated O-glycans, sialylated O-glycans, and both sulfated and sialylated O-glycans, respectively. Among acidic O-glycans, sulfated and sialylated O-glycans showed a particularly strong reduction in the MIX group; whereas the KGM group affected negatively sialylated O-glycans. The abundance of S1F2H2N2 (m/z = 1121) in colonic mucins of obese mice was reported to decrease following supplementation with insoluble NutriKane and Benefiber12. Here, instead, S1F2H2N2 showed an increase, which might be explained by differences in mouse models and type of fiber.
Effect of dietary polysaccharides with different structures on gut microbiota
The influence of structurally diverse polysaccharides on fecal gut microorganisms was demonstrated using 16S rRNA gene amplicon sequencing. The ordination of the gut microbiota in all groups was significant (P < 0.05) by principal component analysis (PCA) and Non-metric multidimensional scaling (nMDS plot) as shown in Supplementary Fig. 5 (includes additional analyses of β-diversity using PERMANOVA, Con vs. LBP: P = 0.019; Con vs. CS: P = 0.027; Con vs. BSP: P = 0.046; Con vs. PEC: P = 0.003; Con vs. KGM: P = 0.007; Con vs. MIX: P = 0.03; Con vs. INU: P = 0.03). Animals supplemented with PEC and MIX displayed higher ɑ-diversity and richness than the CON group or those fed other dietary polysaccharides (Fig. 5A, B). The major phyla were Firmicutes, Bacteroidetes, Verrucomicrobiota, and Proteobacteria, which together represented about 90% of taxa (Fig. 5C). Compared to the CON group, dietary polysaccharides significantly reduced the relative abundance of Proteobacteria (P < 0.05). The PEC group resulted in significantly more Verrucomicrobiota, which were also enriched in the MIX and LBP-AG groups along with Bacteroidetes. The latter were also abundant in the CS and BSP groups.
A, B Diversity indices: Shannon index (A) and ACE index (B). C, D Relative abundance of bacteria at phylum (C) and family (D) level; only abundances >1% are shown. E–G SCFA content in the caecum (mean ± SD): acetic acid (E), propionic acid (F), and butyric acid (G). H Spearman correlation between SCFAs and gut bacteria. Significance (P < 0.05) was determined by Duncan’s test (n = 6).
Comparison at the family (Fig. 5D) and operational taxonomic unit (OTU) level (Fig. 6 and Supplementary Figs. 6 and 7) using a linear discriminant analysis (LDA) effect size (LEfSe) method (LDA > 3) revealed fewer Pseudomonadaceae (OTU 4) following supplementation with dietary polysaccharides. The MIX group yielded significantly (P < 0.05) more OTUs in the family Lachnospiraceae (OTUs 1380, 102, 75, 21, and 133) compared to the CON or other polysaccharide groups. The relative abundance of two OTUs of the genus Akkermansia (OTUs 1 and 1208) was significantly (P < 0.05) higher after supplementation with arabinose-rich PEC, LBP-AG, and MIX. This finding confirms a previous report, whereby a polysaccharide containing arabinose correlated positively with the relative abundance of Akkermansia34. OTUs of the Muribaculaceae were increased in the CS (OTUs 28, 8, 43, 30, 23, 44, 26, 35, 25, and 38) and BSP (OTUs 28, 8, 43, 30, 23, 33, 79, and 44) groups rich in sulfated polysaccharides (Fig. 6). Three OTUs in the genera Bifidobacterium (OTU 15) and Lactobacillus (OTUs 7 and 74) were higher in the KGM and INU groups. The prebiotic INU was previously reported to improve the relative abundance of Lactobacillus and Bifidobacterium7,28, which is consistent with our results. Overall, the various dietary polysaccharides had specific effects on intestinal microbial taxa. Besides OTUs, the contents of butyrate, propionate, and acetate were also assessed in the different groups of mice (Fig. 5E–G). The PEC and MIX groups significantly (P < 0.05) enhanced butyrate content, which was 2.43 times and 2.35 times higher than in the CON group, respectively. Propionate and acetate levels were 1.72 times and 1.45 times greater in the LBP-AG than in the CON group, respectively. Spearman correlation revealed a significant association between SCFA content and specific intestinal bacterial OTUs (Fig. 5H and Supplementary Table 4). The relative abundance of Akkermansia (OTUs 1 and 1208), Lachnospiraceae_NK4A136_group (OTU 2363), and Peptococcaceae (OTU 187) correlated positively with the concentration of butyric acid. Therefore, the increase in butyric acid content in the PEC and MIX groups might be related to enrichment with Akkermansia and Lachnospiraceae_NK4A136_group. Indeed, Lachnospiraceae_NK4A136_group has been shown to promote the release of butyrate35. Propionate content was positively related to the abundance of OTUs in the genera Lachnoclostridium (OTU 102) and GCA-900066575 (OTU 1308).
Differentially abundant OTUs were determined through LDA analyses (LDA > 3) between CON vs. LBP, CON vs. CS, CON vs. BSP, CON vs. PEC, CON vs. KGM, CON vs. MIX, and CON vs. INU (the LDA image and heatmap of relative abundance for each mouse were provided in Supplementary Figs. 6 and 7). The taxonomic level of OTUs (phylum, family, and species) is indicated on the right. Note: “*” (Black) Greater abundance in LBP, CS, BSP, PEC, KGM, MIX, and INU groups compared to CON; “*” (Green) Lower abundance in LBP, CS, BSP, PEC, KGM, MIX, and INU groups compared to CON.
Spearman correlation revealed the relationship between O-glycans and bacterial OTUs (Fig. 7). The relative abundance of OTUs within the genus Bacteroides (OTU 70) correlated positively with the content of non-fucosylated O-glycans such as H1N2 (Number 3). Enrichment of OTUs from Lactobacillus (OTU 74) and Muribaculaceae (OTU 3) correlated positively with the abundance of the H2N2 glycoform (Number 5). F1H1N1 (Number 2) correlated positively with OTUs in the families Prevotellaceae (OTU 25), Muribaculaceae (OTUs 10, 8, 43, 23, 44, and 96), and Oscillospiraceae (OTUs 991, 40, 2, 1064, and 928); but negatively with OTUs in the Lactobacillaceae (OTU 7). Parabacteroides, Lachnoclostridium, and other bacteria stimulate the expression of α-1,2 fucosyltransferase in healthy human feces36, thereby promoting the production of fucosylated O-glycans on intestinal mucin, although this is not observed in patients with colitis37. The difference in microbial composition between this and previous studies may be related to distinct gut microorganisms in human vs. mouse feces. Furthermore, sulfated O-glycan structures such as S1H2N2 (Number 26) correlated negatively with the relative abundance of OTUs in the genus Akkermansia. Instead, S1F2H2N2 (Number 36) O-glycans correlated positively with the relative abundance of OTUs in Lachnospiraceae (OTU 5), Muribaculaceae (OTUs 43, 8, 23, and 44), and Oscillospiraceae (OTUs 991, 40, 56, and 2). Acidic O-glycans containing Neu5Ac are more abundant in the proximal part of the mouse colon, whereas fucosylated O-glycans tend to be more abundant in the distal colon, this distribution contrasts with that in humans, whereby fucosylated O-glycans predominate in the proximal gastrointestinal tract and more acidic O-glycans in the distal one38. In line with our results, sulfated and sialylated O-glycans in mouse colonic Muc2 were reported to correlate positively with OTUs in Muribaculaceae, Lachnospiraceae, and Clostridiales12. In addition, sialylated O-glycans F1H3N2A1 and H2N4A1 exhibited a positive correlation with OTUs within the families Oscillospiraceae (OTUs 19 and 932) and Bacteroidaceae (OTU 1643).
Discussion
In this study, 54 O-glycans were detected in colonic Muc2 of healthy mice. This value exceeds the 37 or 36 O-glycans reported previously12, and might be explained by the separate detection of neutral and acidic O-glycans using positive and negative ion modes to improve MS sensitivity. Tardy et al.39 reported that dietary polysaccharides significantly enhanced the abundance of total and neutral O-glycans on Muc2 in the large intestine of rats by gas chromatography (GC) analysis of glycoprotein sugars. Our results demonstrate that dietary polysaccharides increased the amount of total Muc2 O-glycans and neutral O-glycans using LC-MS, which could be ascribed mainly to an increase in non-fucosylated O-glycans.
Both the PEC and MIX groups significantly increased the expression of neutral fucosylated O-glycans in mouse colonic mucin; whereas the MIX group significantly decreased the content of sulfated and sialylated O-glycans. This disparity might be related to monosaccharide composition and the abundance of glycosidic bonds. The MIX group presented a richer monosaccharide composition and greater glycosidic bond variation than the acidic PEC group, in which the predominant species was α-1,4 galacturonic acid. Hence, the modulatory effect of dietary polysaccharides on O-glycans was related to monosaccharide and glycosidic bond heterogeneity. To this end, corn-resistant starch (containing α-1,4 glucose linkages) has been shown to significantly boost the level of fucosylated O-glycans39. PEC is characterized by the same α-1,4 linkages, suggesting that the presence of fucosylated O-glycans on mucin might be affected by α-1,4 glycosidic linkages. LBP, a highly branched structure characterized by a main chain of β-1,6-Gal units and mostly arabinose substitutions at the O-3 position, significantly increased the content of terminally mono-fucosylated core 1 O-glycans. PEC, a linear heteropolysaccharide harboring α-1,4 glycosidic linkages in the main chain, KGM, a linear glucomannan with β-1,4 linkages, and INU, a linear oligofructose with α-1,2 linkages, boosted the levels of di-fucosylated O-glycans. Therefore, highly branched dietary polysaccharides appear to stimulate mono-fucosylated core 1 O-glycan chains; whereas linear dietary polysaccharides seem to favor di-fucosylated core 2 O-glycan chains. CS, which consists of a repeating disaccharide unit composed of β-1,3-linked glucuronide and GalNAc, as well as PEC, significantly decreased the level of mono-fucosylated O-glycans such as F1H1N2. This result indicates that glyoxylate-based dietary polysaccharides could specifically induce terminally mono-fucosylated core 2 O-glycans. In general, supplementation with PEC and MIX significantly enhanced the content of fucosylated O-glycans, but MIX elicited a stronger reduction in sulfated and sialylated O-glycans. Accordingly, supplementation with a varied mixture of monosaccharides containing different glycosidic bonds had a synergistic and more beneficial effect compared to that triggered by single dietary polysaccharides.
Colonization by specific intestinal bacteria influences O-glycosylation of mucin. For example, oral administration of the probiotic L. farciminis significantly reduced the expression of intestinal sialylated O-glycans (m/z 1331 and 1605) in irritable bowel syndrome18. Furthermore, intestinal bacteria affect glycosyltransferase activity, which in turn alters the degree of O-glycosylation on mucin. Our results demonstrate that differently structured dietary polysaccharides could regulate specific intestinal bacteria. PEC caused a significant rise in the relative abundance of Bacteroides, Lachnospiraceae, and A. muciniphila, following a shift from neutral non-fucosylated O-glycans to H2N2 and F2H2N2. LBP-AG boosted the relative abundance of Prevotellaceae, Muribaculaceae, Oscillospiraceae, and A. muciniphila by favoring mono-fucosylated neutral O-glycans such as F1H1N1. KGM promoted OTUs associated with Bifidobacterium, Lactobacillus, and Muribaculaceae, while decreasing the level of sialylated O-glycans such as F1H3N2A1. In addition, Spearman analysis revealed positive correlations between non-fucosylated O-glycans (H1N2 and H2N2, Numbers 3 and 5) and OTUs from Bacteroides, Lactobacillus, and Muribaculaceae; as well as between the mono-fucosylated O-glycan F1H1N1 (Number 2) and OTUs from the Prevotellaceae, Muribaculaceae, Oscillospiraceae, Lachnospiraceae, and Lactobacillaceae families. In contrast, negative correlations were found between sulfated O-glycans (S1H2N2 and S1F2H2N2, Numbers 26 and 36) and OTUs associated with A. muciniphila, Lachnospiraceae, Muribaculaceae, and Oscillospiraceae; as well as between sialylated O-glycans (F1H3N2A1 and H2N4A1, Numbers 45 and 47) and OTUs in the Oscillospiraceae and Bacteroidaceae families. Therefore, differentially structured dietary polysaccharides might regulate the expression of mouse colonic mucin O-glycans by affecting the composition of intestinal bacteria, and vice versa.
Free monosaccharides/oligosaccharides or SCFAs produced by fermentation of dietary polysaccharides indirectly alter the O-glycosylation pattern of intestinal mucins17,22,23. Bacteroides not only increase the expression of Fut2 in mouse intestinal colonic goblet cells, but they also generate a variety of dietary polysaccharide hydrolases, such as GH2, GH35, and GH42. These, in turn, could degrade mono- and oligosaccharides, including Gal, rhamnose, and arabinose, in the pectin structure and release them into the intestine23. Free Gal might be converted to UDP-Gal via the Leloir pathway to be used as a substrate for the glycosylation of the α1,2Galβ1,3GalNAcol intestinal mucin O-glycan core 2 structure40. GH3, GH43, GH13, GH2, and GH36 hydrolases produced by the Lachnospiraceae family metabolize butyrate and have been shown to improve colonic mucin16,41,42. The latter was found to increase the relative abundance of A. muciniphila, which used mucin O-glycans as a carbon source43. Our results revealed that the intestinal bacteria of mice in the PEC group were dominated by Bacteroides, Lachnospiraceae_NK4A136_group, and A. muciniphila. The same group was characterized by significantly higher levels of total O-glycans and neutral fucosylated O-glycans, which might be attributed to upregulation Fut2 expression due to an increased abundance of Bacteroides-related OTUs. Meanwhile, PEC could also enhance the expression of O-glycans through interactions of Lachnospiraceae and A. muciniphila. Prevotellaceae encodes specific trsusC/D polysaccharide utilization sites that produce hydrolases (e.g., GH137 and xynD)44, degrade dietary polysaccharides into free monosaccharide or oligosaccharide units, and release acetate, which promotes KLF4 signaling and consequently inhibits neutral O-glycans in mouse colonic goblet cells17. Our results point to a significantly higher relative abundance of Prevotellaceae and acetate content in the LBP-AG, CS, and BSP groups compared to other dietary polysaccharide groups; but a lower content of neutral O-glycans. We hypothesize that this observation might be due to LBP-AG, CS, and BSP being able to increase acetate levels by stimulating the growth of Prevotellaceae and consequent downregulation of neutral O-glycan chains in mouse intestinal mucins. The O-glycan profile was similar for INU (composed of β-1,2-linked fructose) and KGM (composed of glucomannan linked by β-1,4 glycosidic bonds), and was characterized by fewer terminally mono-fucosylated and sialylated O-glycans. This similarity might be explained by the degradation of free mannose by intestinal microorganisms (Bifidobacterium and Lactobacillus)45, as well as its isomerization to fructose via phosphomannose isomerase. The impact of structurally diverse dietary polysaccharides on intestinal bacteria or colonization with specific intestinal microorganisms on the O-glycans of intestinal mucins has been reported previously18,23.
In this study, we investigated changes to the intestinal microbial composition and its metabolites (e.g., SCFAs), as well as the O-glycans of colonic mucins following the administration of six structurally defined dietary polysaccharides. The correlation between intestinal microbes and colonic mucin O-glycans was analyzed using bioinformatics, and the possible relationship between dietary polysaccharide structure, intestinal flora, and colonic mucin O-glycans was explored. These correlations could help clarify the relationship between colonic mucin O-glycans and intestinal microbiota in both healthy and diseased individuals.
In summary, dietary polysaccharides maintain a healthy gut by influencing the colonic mucus layer of highly O-glycosylated mucins. This is the first study to elucidate how intestinal mucin O-glycosylation is influenced by polysaccharide structure. Highly branched arabinogalactan favored terminally fucosylated core 1 O-glycans; whereas linear chain polysaccharides, including pectin, konjac glucomannan, oligofructose (inulin), and a polysaccharide mixture, increased significantly the abundance of terminally di-fucosylated O-glycans. In addition, the relationship between dietary polysaccharide structure, mucin O-glycan composition, and intestinal microorganisms was systematically compared, opening the door for targeted nutritional interventions.
Methods
Experimental model and subject details
Animal model
C57BL/6J mice, aged 6–8 weeks and weighing approximately 20.0–22.0 g, were housed under specific pathogen-free conditions. Standard lighting (12-h light/dark cycle), humidity, and temperature (25 ± 2 °C), as well as adequate food and germ-free drinking water (autoclaved) were provided. Animals were randomly assigned to eight groups, with 10 mice per group, after adaptive rearing for a week (Fig. 1B). Daily intake of the main nutrients is reported in Fig. 1C, and diet composition is reported in Supplementary Table 2. The different polysaccharides (LBP, CS, BSP, PEC, KGM, INU and their mixture) were dissolved in saline (0.9% NaCl), and gavaged daily (200 mg/kg/day) to mice for 9 weeks, the mixture group (MIX, 200 mg) was prepared six polysaccharide mixture with an equal amount; meanwhile, the CON group was orally administrated with an equal volume of saline. Body weight and feed quantity were recorded daily. During the last 3 days of the trial, each mouse was placed in the metabolism cage (LAT- XSDXL, Lab Animal Technology Develop Co., Beijing, China), respectively, and fecal samples were collected into sterile centrifuge tubes (1.5 mL) using tweezers and fast-frozen in liquid nitrogen for 16S rRNA sequencing. The mice were euthanized with isoflurane after 9 weeks of the dietary intervention, at which point their blood, cecal contents, cecum, colon, kidney, spleen, and liver were collected, weighted, and stored at –80 °C. All animal studies (including the mice euthanasia procedure) were approved by Experimental Animal Management and ethics guidelines of Northwest University in China (NWU-AWC-20210401M). The approval declaration is provided in Supplemental Information.
Polysaccharide extraction and determination of basic indices
For LBP-AG extraction, the fruits were crushed and extracted twice at 80 °C for 2 h, with the two resulting supernatants pooled together and concentrated at <45 °C. Next, the concentrated supernatants were precipitated for 12 h using ethanol, and the precipitates were dissolved in Sevage reagent (1:4 n-butanol:chloroform) to remove proteins. Finally, the samples were dialyzed for 3 days, concentrated, and precipitated with a graded ethanol series (30%, 50%, and 70%). The 70% precipitates were vacuum freeze-dried to obtain LBP-AG. KGM and CS were purchased from Hebei Bailingway Superfine Materials Co Ltd (Hebei, China), INU was bought from Aladdin (Shanghai, China), while BSP and PEC were obtained from Meiyichen Biotechnology Company (Jiangsu, China). KGM, CS, INU, BSP, and PEC were dialyzed (3000 kDa) and freeze-dried. Sugar content was detected with the phenol sulfuric acid method. Protein content was measured with a BCA kit (Beyotime Biotechnology, Nanjing, China). The monosaccharide components and molecular weight of different polysaccharides were measured as reported previously26. Protein and sugar content, monosaccharide composition, and molecular weights of polysaccharides are listed in Supplementary Table 1.
Colon pathological sections
Colons were rinsed with pre-chilled phosphate-buffered saline and were fixed using Carnoy’s fixative (n = 6). The colon tissue specimens were embedded in paraffin after dehydration with ethanol, sliced into sections of 2–3 µm, deparaffinized, and treated with periodic acid alcohol solution and Schiff reagent, as well as hematoxylin and eosin. After that, the samples were dehydrated and sealed. In addition, sections were placed in Alcian Blue Stain Solution A for 15 min, washed with water, stained with Alcian Blue Stain Solution B for 3 min, dehydrated, and sealed. Histological sections were photographed at ×4, ×10, and ×20 magnification using an inverted fluorescence microscope (ECLIPSE Ti2-A; Nikon, Tokyo, Japan). For magnifications above ×10, three random fields of view were imaged. In addition, the number of goblet cells and crypts (×4 magnification PAS) was determined using ImageJ version 1.51j8 (NIH, Bethesda, MD, USA).
Immunofluorescence staining of colonic mucin 2
The colon (1.0 cm) was fixed using Carnoy’s fixative (n = 6). The samples were dehydrated in ethanol, embedded in paraffin, sliced into sections of 2–3 µm, and incubated in the dark with primary antibodies against mucin 2 (1:2000, 27675-1-AP; Proteintech, Wuhan, China) and secondary antibodies (1:3000, G1215-200T; Servicebio, Wuhan, China). Finally, the slides were observed under a confocal microscope (AX-R; Nikon) at ×10, ×20, and ×40 magnification. Different fields of view were selected at random and imaged three times. Mucin 2 (×10 magnification) levels were quantified using ImageJ.
Western blotting
Mouse colon samples of 70–90 mg were weighted and minced (n = 6), before resuspending the resulting lysate in RIPA buffer (PR20001; Proteintech) containing phenylmethylsulfonyl fluoride (PMSF, 20016; Proteintech) as protease inhibitor. The samples were disrupted with an ultrasonicator and the supernatant was collected by centrifugation. Total protein content was assayed with the BCA method (Beyotime Biotechnology). Immunoblotting of β-actin (1:2000, GB1500; Servicebio), GADPH (1:5000, M20050S, Abmart, Shanghai, China), ZO-1 (1:500, GB11140; Servicebio), (1:1000, bs-12258R; Bioss, Beijing, China) occludin (1:500, GB111401, Servicebio), claudin 1 (1:500, GB11254; Servicebio), and Fut2 (1:1000, bs-12258R; Bioss, Beijing, China) was performed as described previously using a fluorescence chemiluminescence imaging system26. Images were processed with ImageJ, with each group assessed at least three times (n ≥ 4).
Sequencing and bioinformatics analysis of 16S rRNA gene amplicons
Briefly, 2–4 g (n = 6) of mouse feces were weighed and total genomic DNA of mouse fecal bacteria was extracted using a QIAamp DNA Stool Mini Kit (51504; QIAGEN, Holden, Germany).
Amplicons were generated as summarized hereafter. Different regions of the 16S rRNA gene (V3-V4) were amplified using specific primers with barcodes (319F/806R). All PCR reactions were performed with 15 μL Phusion® High Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA), 0.2 μM forward and reverse primers, and approximately 10 ng template DNA. Thermal cycling included initial denaturation at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and extension at 72 °C for 30 s, plus a final step of 5 min at 72 °C.
To quantify and identify the PCR products, the same volume of SYBR (1×) loading buffer (containing SYBR Green) was mixed with the PCR products, which were then separated on a 2% agarose gel and purified with the Qiagen Gel Extraction Kit.
Sequencing libraries were generated using TruSeq® DNA PCR-Free sample preparation kits (Illumina, San Diego, CA, USA) according to the manufacturer’s recommendations and index codes were added. The libraries were quantified on a Qubit@2.0 Fluorometer (Thermo Scientific) and evaluated on an Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Finally, libraries were sequenced on the Illumina NovaSeq 6000 platform to generate 250-bp paired-end reads.
Paired reads were assigned to samples based on their unique barcodes, and truncated by cutting off barcodes and primer sequences. They were then combined using FLASH (VI.2.7, http://ccb.jhu.edu/software/FLASH/)46, a very fast and accurate tool designed to merge paired-end reads when at least some of them overlap with reads generated at the other end of the same DNA fragment. The spliced sequence, referred to as the original tag, was quality-filtered under specific filtering conditions following the QIIME V1.9.1 (http://qiime.org/scripts/split_libraries_fastq.html) quality control process to obtain high-quality clean labels47,48. The tags were compared to the reference Silva database (https://www.arb-silva.de/) using the UCHIME algorithm (http://www.drive5.com/usearch/manual/uchime_algo.html) to detect and remove chimeric sequences from the final label49,50. An average of 81,255 tags was measured per sample by splicing the reads, and an average of 80,637 valid data were obtained through quality control, yielding 62,022 valid data and a quality control efficiency of 76.35%. Sequences were clustered into OTUs with 97% identity using Uparse, which yielded a total of 2588 OTUs. Finally, the OTUs were annotated with the Silva138 database for species.
Non-metric multidimensional scaling (nMDS) plots and principal component analysis (PCA) were constructed based on Bray–Curtis similarity matrices of Log (x + 1) transformed abundance of the OTUs, Permutational Multivariate Analysis of Variance (PERMANOVA) tests based on Bray–Curtis distance measures to investigate differences in the microbial community structure, and PERMANOVA were analyzed by Prizer 7.0. Distinct phylotypes (families and OTUs) between dietary groups were identified using the LEfSe method with the following parameters: Kruskal-Wallis test among groups (P < 0.05), Wilcoxon test between groups (P < 0.05), and the threshold on the LDA score for discriminative features >3.051. The correlation was determined using Spearman analysis. Bioinformatics analysis and image was performed using the OmicStudio tools at https://www.omicstudio.cn/tool.
Quantification of SCFAs
SCFA standards were prepared by transferring 9.52 μL acetic acid, 10.10 μL propionic acid, 10.10 μL butyric acid, 10.54 μL isobutyric acid, 10.66 μL n-valeric acid, 10.80 μL isovaleric acid, and 99.38 μL ether to a 15-mL centrifuge tube, which was then vortexed and mixed to obtain a stock solution of the six SCFAs. The stock solution was diluted to different concentrations (5, 50, 100, 200, 300, 400, and 500 mM).
Next, 100 mg of feces were transferred to a 2-mL centrifuge tube, followed by addition of steel beads for stirring, acidification with H2SO4 (50%, v/v), addition of diethyl ether, and vortexing twice. The fecal samples were ultrasonicated for 30 min in an ice bath at –20 °C, mixed with anhydrous sodium sulfate, vortexed, and finally centrifuged at 12,000 × g (15 min, 4 °C). The supernatant was filtered using a 0.22-μm organic filter.
Gas chromatography was carried out on a GC2010-PLUS instrument (Shimadzu, Kyoto, Japan), equipped with an HP-INNOWax column (0.25 μm, 0.250 mm × 30 mm; Agilent Technologies). The following parameters were used: inlet temperature, 250 °C; FID temperature, 230 °C; and split ratio, 10:1. The samples were heated to an initial temperature of 100 °C, ramped to 180 °C at 5 °C/min, and then held at 180 °C for 4 min. The separation took a total of 21 min, with a makeup of 9 min using air at 400 mL/min, N2 at 3 mL/min, and H2 at 40 mL/min.
Mucin sample collection and O-glycan structure determination
The colon was dissected, scraped with tweezers, and dissolved in 6 M guanidine chloride containing 0.01 M NaH2PO4 and 5 mM EDTA. PMSF (2 mM) was added prior to extraction with a glass mini grinder. After stirring with a pestle at 4 °C overnight, 10 mL of samples were reduced with dithiothreitol and methylate/iodoacetamide, dialyzed with 2 L of distilled water at 4 °C for 3 days (10 kDa), and changed distilled water twice a day at 8:00 am and 8:00 pm, transferred to centrifuge tubes, and freeze-dried.
O-glycans were released adding 1 mL NaOH (0.1 M) and 1 mL NaBH4 (1 M), and incubating for 15 h at 50 °C. β-Cyclodextrin (99.99%) and 6′-sialyllactose (99.9%) were added as internal standards, while acetate was added to adjust the pH to 4.5. The samples were purified using Solid-phase extraction (SPE) of C18 cartridges (200 mg/3 mL, SAC-C1803200; Simon Aldrich, Germany) and graphitic carbon SPE (200 mg/3 mL, Supelco; Bellefonte, PA, USA), A DEAE column (2 mg/3 mL; Biobomei, Hefei, China) was used to separate neutral and acidic O-glycans, which were detected under positive and negative ion modes, respectively (Supplementary Figs. 2 and 3).
O-glycans were analyzed using LC-MS. High-performance liquid chromatography was performed on a 1290 Infinity system (Agilent Technologies) using a Hypercarb column (3 μm, 2.1 mm × 100 mm; Thermo Scientific). Sample injection volume was 5 μL, column temperature was 60 °C, and flow rate was 0.450 mL/min. Mobile phases A (water) and B (1% acetonitrile in 0.1% v/v formic acid) were used to form the gradients as follows: 0–5 min, 100–85% A; 5–15 min, 85–70% A; 15–30 min, 70–60% A; 30–35 min, 60–30% A; 35–40 min, 30–5% A; 40–42 min, 5–2% A; 42–45 min, 2–0% A; and 45–50 min, 0–100% A. In negative ion mode, the program was as follows: 0–8 min, 100–85% A; 8–15 min, 85–75% A; 15–25 min, 75–70% A; 25–35 min, 70–60% A; 35–45 min, 60–40% A; 45–50 min, 40–2% A; 50–60 min, 2–0% A; and 60–65 min, 0–100% A.
LC-MS was carried out on a G6400 quadrupole time-of-flight mass spectrometer (Agilent Technologies) in positive or negative ion modes using a jet stream electrospray ionization source. The source parameters were as follows: sheath gas (N2), 400 °C at 12 L/min; drying gas (N2), 300 °C at 10 L/min; nozzle voltage, 1000 V; capillary voltage, 4750 V; fragment voltage, 175 V; and nebulizer pressure, 45 psi.
LC-MS data acquisition and analysis were performed using MassHunter Workstation software (Agilent Technologies) and Data Acquisition Workstation (v B.06.01, SP1). Peak areas and retention times were processed further in Microsoft Excel. Glycan compositions and structures were presumed according to a combination of the m/z value, regularity of O-glycans synthesis, published literature, and GlycoWorkbench version 2.13,12,32.
Statistical analysis
Statistical analyses were performed using one-way ANOVA with Duncan’s test by GraphPad Prism version 8.0 (GraphPad Inc., La Jolla, CA, USA) unless otherwise stated. For comparisons between two groups, an unpaired t-test was used when the samples were normally distributed. In all figures, data are presented as the mean ± standard error of the mean (SEM) or mean ± standard deviation (SD). Spearman correlations were used for data that were not normally distributed.
Data availability
All data supporting the findings of this study are available within the article and its Supplemental Information files. The non-curated MS raw data (MS/MS of isomer) was updated in UniCarbDR (https://unicarb-dr.glycosmos.org/references). The data of 16S rRNA gene amplicons are available from the NCBI under accession PRJNA1016085. The data generated and analyzed during the present study are available from the corresponding author upon reasonable request.
References
Paone, P. & Cani, P. D. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut 69, 2232–2243 (2020).
Hansson, G. C. Mucins and the Microbiome. Annu. Rev. Biochem. 89, 769–793 (2020).
Arike, L., Holmén-Larsson, J. & Hansson, G. C. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology 27, 318–328 (2017).
Atuma, C., Strugala, V., Allen, A. & Holm, L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 280, G922–G929 (2001).
Bansil, R. & Turner, B. S. The biology of mucus: composition, synthesis and organization. Adv. Drug Deliv. Rev. 124, 3–15 (2018).
Fang, J. et al. Slimy partners: the mucus barrier and gut microbiome in ulcerative colitis. Exp. Mol. Med. 53, 772–787 (2021).
Zhang, D. et al. Interactions between polysaccharides and gut microbiota: a metabolomic and microbial. Food Res. Int. 160, 111653 (2022).
Johansson, M. E. V. & Hansson, G. C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 16, 639–649 (2016).
Larsson, H. et al. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G357–G363 (2013).
An, G. et al. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. Inflamm. Bowel Dis. 13, 654–655 (2007).
Qu, D. et al. The effects of diet and gut microbiota on the regulation of intestinal mucin glycosylation. Carbohydr. Polym. 258, 1–15 (2021).
Gamage, H. K. A. H., Chong, R. W. W., Bucio-Noble, D., Kautto, L. & Paulsen, I. T. Changes in dietary fiber intake in mice reveal associations between colonic mucin O-glycosylation and specific gut bacteria. Gut Microbes 12, e1802209–e1802224 (2020).
Sorbara, M. T. & Pamer, E. G. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal. Immunol. 1–9. https://doi.org/10.1038/s41385-018-0053-0 (2019).
Martin-Gallausiaux, C. et al. Akkermansia muciniphila upregulates genes involved in maintaining the intestinal barrier function via ADP-heptose-dependent activation of the ALPK1/TIFA pathway. Gut Microbes 14, e2110639 (2022).
Kim, Y. S. & Ho, S. B. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr. Gastroenterol. Rep. 12, 319–330 (2010).
Tailford, L. E., Crost, E. H., Kavanaugh, D. & Jug, N. Mucin glycan foraging in the human gut microbiome. Frontiers 6, 00081 (2015).
Wrzosek, L. et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 11, 2–13 (2013).
Silva, S. D. et al. Stress disrupts intestinal mucus barrier in rats via mucin O-glycosylation shift: prevention by a probiotic treatment. Am. J. Physiol. Gastrointest. Liver Physiol. 307, 420–429 (2014).
Johansson, M. E. et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 18, 582–592 (2015).
Duan, H., Wang, L., Huangfu, M. & Li, H. The impact of microbiota-derived short-chain fatty acids on macrophage activities in disease: mechanisms and therapeutic potentials. Biomed. Pharmacother. 165, 115276 (2023).
Hasnain, S. Z. et al. IL-10 promotes production of intestinal mucus by suppressing protein misfolding and endoplasmic reticulum stress in goblet cells. Gastroenterology 144, 357–368 (2013).
David, A. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Tuncil, Y. E., Thakkar, R. D., Arioglu-Tuncil, S., Hamaker, B. R. & Lindemann, S. R. Subtle variations in dietary-fiber fine structure differentially influence the composition and metabolic function of gut microbiota. mSphere 5, 00180–000120 (2020).
Mao, G. et al. Depolymerized RG-I-enriched pectic polysaccharide from citrus segment membranes modulates gut microbiota, increases SCFA production, and promotes the growth of Bifidobacterium spp., Lactobacillus spp. and Faecalibaculum spp. Food Funct. 10, 7828–7843 (2019).
Larsen, N. et al. Potential of pectins to beneficially modulate the gut microbiota depends on their structural properties. Frontiers 10, 223 (2019).
Cao, C. et al. An arabinogalactan from Lycium barbarum attenuates DSS-induced chronic colitis in C57BL/6J mice associated with the modulation of intestinal barrier function and gut microbiota. Food Funct. 12, 9829–9843 (2021).
Kang, Y., Xue, Y., Du, M. & Zhu, M. J. Preventive effects of Goji berry on dextran-sulfate-sodium-induced colitis in mice. J. Nutr. Biochem. 40, 70–76 (2017).
Changchien, C.-H., Han, Y.-C. & Chen, H.-L. Konjac glucomannan polysaccharide and inulin oligosaccharide enhance the colonic mucosal barrier function and modulate gut-associated lymphoid tissue immunity in C57BL/6J mice. Br. J. Nutr. 123, 319–327 (2020).
Wu, D. et al. Protective effects of six different pectic polysaccharides on DSS-induced IBD in mice. Food Hydrocoll. 127, 1–11 (2021).
Shimotoyodome, A., Mecuro, S., Hase, T., Tokimitsu, I. & Sakata, T. Sulfated polysaccharides, but not cellulose, Increase colonic mucus in rats with loperamide-Induced constipation. Digestive Dis. Sci. 46, 1482–1489 (2001).
Xie, S.-Z. et al. Dendrobium huoshanense polysaccharide regionally regulates intestinal mucosal barrier function and intestinal microbiota in mice. Carbohydr. Polym. 206, 149–162 (2019).
Bergstrom, K. et al. Proximal colon–derived O-glycosylated mucus encapsulates and modulates the microbiota. Science https://doi.org/10.1126/science.aay7367 (2020).
Sharma, R., Schumacher, U., Ronaasen, V. & Coates, M. Rat intestinal mucosal responses to a microbial flora and different diets. Gut 36, 209–214 (1995).
Calvete-Torre, I. et al. Prebiotic potential of apple pomace and pectins from different apple varieties: Modulatory effects on key target commensal microbial populations. Food Hydrocoll. 133, 1–13 (2022).
Pereira, M. D. F. A. et al. The role of IL-10 in regulating inflammation and gut microbiome in mice consuming milk kefir and orally challenged with S. Typhimurium. Food Funct. 14, 3804–3814 (2023).
Cheng, S. et al. Altered gut microbiome in Fut2 loss-of-function mutants in support of personalized medicine for inflammatory bowel diseases. J. Genet. Genomics 48, 771–780 (2021).
Kang, S. et al. The impact of Fut2 Loss-of-Function mutation on the pathogenesis of inflammatory bowel diseases mediated by the gut microbiome. Gastroenterology 160, S-119 (2015).
Robbe, C. et al. Evidence of regio-specific glycosylation in human intestinal mucins: presence of an acidic gradient along the intestinal tract. J. Biol. Chem. 278, 46337–46348 (2003).
Tardy, F., Louisot, P. & Martin, A. Effect of dietary fiber at weaning on protein glycosylation in the rat small intestine. Int. J. Biochem. Cell Biol. 27, 403–413 (1995).
Csiszovszki, Z., Krishna, S., Orosz, L., Adhya, S. & Semsey, S. Structure and function of the d-Galactose network in enterobacteria. mBio 2, e00053–00011 (2011).
Bang, S.-J. et al. The influence of in vitro pectin fermentation on the human fecal microbiome. AMB Express 8, 2–9 (2018).
Gaudier, E., Rival, M., Buisine, M. P., Robineau, I. & Hoebler, C. Butyrate enemas upregulate Muc genes expression but decrease adherent mucus thickness in mice colon. Physiol. Res. 58, 111–119 (2009).
Shuoker, B. et al. Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria. Nat. Commun. 14, 1833 (2023).
Gálvez, E. J. C. et al. Distinct polysaccharide utilization determines interspecies competition between intestinal prevotella spp. Cell Host Microbe 28, 838–852 (2020).
Wang, P. et al. Recent advances in biotransformation, extraction and green production of D-mannose. Curr. Res. Food Sci. 5, 49–56 (2022).
Magoc, T. & Salzberg, S. L. FLASH:fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Bokulich, N. A. et al. Quality-filtering vastly improves diversity estimates from Illuminaamplicon sequencing. Nat. Methods 10, 57–59 (2013).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011).
Haas, B. J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504 (2011).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Acknowledgements
This research was supported by the National Natural Science Foundation of China (grant numbers: 31901694, 32171278, and 31972024) and Shaanxi Province Innovation Capability Support Plan-Science and Technology Innovation Team (2020TD-044).
Author information
Authors and Affiliations
Contributions
T.Z. and G.G.: data curation. T.Z.: writing—review and editing. T.Z.: writing—original draft. Zh W.: visualization. Y.Zh., L.L., X.D., Y.L., and X.L.: investigation. G.G., Y.Zh., L.N., Y.X. and S.W.: data acquisition. Q.Zh.: conceptualization. C.C.: writing—review. G.G.: funding acquisition. Zh.W., G.G. and L.H.: supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, T., Zhang, Y., Nan, L. et al. Impact of structurally diverse polysaccharides on colonic mucin O-glycosylation and gut microbiota. npj Biofilms Microbiomes 9, 97 (2023). https://doi.org/10.1038/s41522-023-00468-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-023-00468-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.