Abstract
Preeclampsia and gestational hypertension are common pregnancy complications associated with adverse maternal and child outcomes. Current tools for prediction, prevention and treatment are limited. Here we tested the association of maternal DNA sequence variants with preeclampsia in 20,064 cases and 703,117 control individuals and with gestational hypertension in 11,027 cases and 412,788 control individuals across discovery and follow-up cohorts using multi-ancestry meta-analysis. Altogether, we identified 18 independent loci associated with preeclampsia/eclampsia and/or gestational hypertension, 12 of which are new (for example, MTHFR–CLCN6, WNT3A, NPR3, PGR and RGL3), including two loci (PLCE1 and FURIN) identified in the multitrait analysis. Identified loci highlight the role of natriuretic peptide signaling, angiogenesis, renal glomerular function, trophoblast development and immune dysregulation. We derived genome-wide polygenic risk scores that predicted preeclampsia/eclampsia and gestational hypertension in external cohorts, independent of clinical risk factors, and reclassified eligibility for low-dose aspirin to prevent preeclampsia. Collectively, these findings provide mechanistic insights into the hypertensive disorders of pregnancy and have the potential to advance pregnancy risk stratification.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
GWAS summary statistics for preeclampsia/eclampsia and gestational hypertension and genome-wide polygenic scores for preeclampsia/eclampsia, gestational hypertension and systolic blood pressure are available for download at https://doi.org/10.6084/m9.figshare.22680904.v1. Polygenic scores are also available in the PGS Catalog (https://www.pgscatalog.org/publication/PGP000462/). Summary statistics used in this meta-analysis are publicly available for FinnGen r6 (https://www.finngen.fi/en/access_results) and for BioBank Japan (https://pheweb.jp/pheno/PreEclampsia). Preeclampsia GWAS summary statistics from the InterPregGen consortium are available at https://ega-archive.org (dataset IDs EGAD00010001984 (European maternal meta-analysis), EGAD00010001985 (Central Asian maternal meta-analysis) and EGAD00010001983 (European and Central Asian fetal meta-analysis)). Placental transcriptome data are publicly available at https://www.obgyn.cam.ac.uk/placentome/.
Code availability
The code used to conduct these analyses is available at https://github.com/buutrg/HDP.
References
Burton, G. J., Redman, C. W., Roberts, J. M. & Moffett, A. Pre-eclampsia: pathophysiology and clinical implications. BMJ 366, l2381 (2019).
Jiang, L. et al. A global view of hypertensive disorders and diabetes mellitus during pregnancy. Nat. Rev. Endocrinol. 18, 760–775 (2022).
Garovic, V. D. et al. Incidence and long-term outcomes of hypertensive disorders of pregnancy. J. Am. Coll. Cardiol. 75, 2323–2334 (2020).
Magee, L. A. et al. The 2021 International Society for the Study of Hypertension in pregnancy classification, diagnosis & management recommendations for international practice. Pregnancy Hypertens. 27, 148–169 (2022).
ACOG practice bulletin no. 202: gestational hypertension and preeclampsia. Obstet. Gynecol. 133, 1 (2019).
Honigberg, M. C. et al. Long-term cardiovascular risk in women with hypertension during pregnancy. J. Am. Coll. Cardiol. 74, 2743–2754 (2019).
Rana, S., Lemoine, E., Granger, J. & Karumanchi, S. A. Preeclampsia: pathophysiology, challenges, and perspectives. Circ. Res. 124, 1094–1112 (2019).
Levine, R. J. et al. Circulating angiogenic factors and the risk of preeclampsia. N. Engl. J. Med. 350, 672–683 (2004).
Bartsch, E., Medcalf, K. E., Park, A. L. & Ray, J. G. Clinical risk factors for pre-eclampsia determined in early pregnancy: systematic review and meta-analysis of large cohort studies. BMJ 353, I1753 (2016).
Cnattingius, S., Reilly, M., Pawitan, Y. & Lichtenstein, P. Maternal and fetal genetic factors account for most of familial aggregation of preeclampsia: a population-based Swedish cohort study. Am. J. Med. Genet. A 130, 365–371 (2004).
Nilsson, E., Salonen Ros, H., Cnattingius, S. & Lichtenstein, P. The importance of genetic and environmental effects for pre-eclampsia and gestational hypertension: a family study. BJOG 111, 200–206 (2004).
McGinnis, R. et al. Variants in the fetal genome near FLT1 are associated with risk of preeclampsia. Nat. Genet. 49, 1255–1260 (2017).
Steinthorsdottir, V. et al. Genetic predisposition to hypertension is associated with preeclampsia in European and Central Asian women. Nat. Commun. 11, 5976 (2020).
Honigberg, M. C. et al. Genetic variation in cardiometabolic traits and medication targets and the risk of hypertensive disorders of pregnancy. Circulation 142, 711–713 (2020).
Gray, K. J. et al. Risk of pre-eclampsia in patients with a maternal genetic predisposition to common medical conditions: a case-control study. BJOG 128, 55–65 (2021).
O’Kelly, A. C. et al. Pregnancy and reproductive risk factors for cardiovascular disease in women. Circ. Res. 130, 652–672 (2022).
Kivioja, A. et al. Increased risk of preeclampsia in women with a genetic predisposition to elevated blood pressure. Hypertension 79, 2008–2015 (2022).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Giri, A. et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat. Genet. 51, 51–62 (2019).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Turley, P. et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 50, 229–237 (2018).
Padmanabhan, S., Caulfield, M. & Dominiczak, A. F. Genetic and molecular aspects of hypertension. Circ. Res. 116, 937–959 (2015).
Rubattu, S., Forte, M., Marchitti, S. & Volpe, M. Molecular implications of natriuretic peptides in the protection from hypertension and target organ damage development. Int. J. Mol. Sci. 20, 798 (2019).
Ohwaki, A. et al. Altered serum soluble furin and prorenin receptor levels in pregnancies with pre-eclampsia and fetal growth restriction. J. Gynecol. Obstet. Hum. Reprod. 50, 102198 (2021).
Battle, A. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Ghoussaini, M. et al. Open targets genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 49, D1311–D1320 (2021).
Weeks, E. M. et al. Leveraging polygenic enrichments of gene features to predict genes underlying complex traits and diseases. Preprint at medRxiv https://doi.org/10.1101/2020.09.08.20190561 (2020).
Gong, S. et al. The RNA landscape of the human placenta in health and disease. Nat. Commun. 12, 2639 (2021).
Maynard, S. E. et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J. Clin. Invest. 111, 649–658 (2003).
Tekola-Ayele, F. et al. Placental multi-omics integration identifies candidate functional genes for birthweight. Nat. Commun. 13, 2384 (2022).
Bai, X. et al. The smooth muscle-selective RhoGAP GRAF3 is a critical regulator of vascular tone and hypertension. Nat. Commun. 4, 2910 (2013).
Kalluri, A. S. et al. Single-cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation 140, 147–163 (2019).
Ge, T., Chen, C. Y., Ni, Y., Feng, Y. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Davidson, K. W. et al. Aspirin use to prevent preeclampsia and related morbidity and mortality: US preventive services task force recommendation statement. JAMA 326, 1186–1191 (2021).
Pollheimer, J. et al. Activation of the canonical wingless/T-cell factor signaling pathway promotes invasive differentiation of human trophoblast. Am. J. Pathol. 168, 1134–1147 (2006).
Zhang, Z. et al. Wnt/β-catenin signaling pathway in trophoblasts and abnormal activation in preeclampsia (review). Mol. Med. Rep. 16, 1007–1013 (2017).
Tita, A. T. et al. Treatment for mild chronic hypertension during pregnancy. N. Engl. J. Med. 386, 1781–1792 (2022).
Zhang, W. et al. Atrial natriuretic peptide promotes uterine decidualization and a TRAIL-dependent mechanism in spiral artery remodeling. J. Clin. Invest. 131, e151053 (2021).
Maack, T. et al. Physiological role of silent receptors of atrial natriuretic factor. Science 238, 675–678 (1987).
Gu, Y. et al. Aberrant pro-atrial natriuretic peptide/corin/natriuretic peptide receptor signaling is present in maternal vascular endothelium in preeclampsia. Pregnancy Hypertens. 11, 1–6 (2018).
Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018).
Hauspurg, A. et al. Association of N-terminal pro-brain natriuretic peptide concentration in early pregnancy with development of hypertensive disorders of pregnancy and future hypertension. JAMA Cardiol. 7, 268–276 (2022).
Satpathy, A. T. et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209, 1135–1152 (2012).
Wang, Y. et al. ZBTB46 is a shear-sensitive transcription factor inhibiting endothelial cell proliferation via gene expression regulation of cell cycle proteins. Lab. Invest. 99, 305–318 (2019).
Hall, G., Wang, L. & Spurney, R. F. TRPC channels in proteinuric kidney diseases. Cells 9, 44 (2019).
Wang, Z. et al. Transient receptor potential cation channel 6 contributes to kidney injury induced by diabetes and hypertension. Am. J. Physiol. Renal Physiol. 322, F76–F88 (2022).
Ives, C. W., Sinkey, R., Rajapreyar, I., Tita, A. T. N. & Oparil, S. Preeclampsia-pathophysiology and clinical presentations: JACC state-of-the-art review. J. Am. Coll. Cardiol. 76, 1690–1702 (2020).
Wang, W. et al. LNK/SH2B3 loss of function promotes atherosclerosis and thrombosis. Circ. Res. 119, e91–e103 (2016).
Machiela, M. J. & Chanock, S. J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015).
Deloukas, P. et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 45, 25–33 (2013).
Gupta, A. K., Hasler, P., Holzgreve, W. & Hahn, S. Neutrophil NETs: a novel contributor to preeclampsia-associated placental hypoxia? Semin. Immunopathol. 29, 163–167 (2007).
Dou, H. et al. Oxidized phospholipids promote netosis and arterial thrombosis in LNK(SH2B3) deficiency. Circulation 144, 1940–1954 (2021).
Wright, D., Syngelaki, A., Akolekar, R., Poon, L. C. & Nicolaides, K. H. Competing risks model in screening for preeclampsia by maternal characteristics and medical history. Am. J. Obstet. Gynecol. 213, e1–e10 (2015).
Akolekar, R., Syngelaki, A., Poon, L., Wright, D. & Nicolaides, K. H. Competing risks model in early screening for preeclampsia by biophysical and biochemical markers. Fetal Diagn. Ther. 33, 8–15 (2013).
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019).
Angras, K. et al. Retrospective application of algorithms to improve identification of pregnancy outcomes from the electronic health record. J. Perinatol. 43, 10–14 (2023).
Klungsøyr, K. et al. Validity of pre-eclampsia registration in the medical birth registry of Norway for women participating in the Norwegian mother and child cohort study, 1999–2010. Paediatr. Perinat. Epidemiol. 28, 362–371 (2014).
Klemmensen, A. K., Olsen, S. F., Osterdal, M. L. & Tabor, A. Validity of preeclampsia-related diagnoses recorded in a national hospital registry and in a postpartum interview of the women. Am. J. Epidemiol. 166, 117–124 (2007).
Kurki, M. I. et al. FinnGen provide genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023).
Sun, B. B. et al. Genetic associations of protein-coding variants in human disease. Nature 603, 95–102 (2022).
Zhou, W. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018).
Leitsalu, L. et al. Cohort profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int. J. Epidemiol. 44, 1137–1147 (2015).
Finer, S. et al. Cohort profile: East London Genes & Health (ELGH), a community-based population genomics and health study in British Bangladeshi and British Pakistani people. Int. J. Epidemiol. 49, 20–21 (2020).
Wei, W. Q. et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS ONE 12, e0175508 (2017).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021).
Sakaue, S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53, 1415–1424 (2021).
Honigberg, M. C. et al. Heart failure in women with hypertensive disorders of pregnancy: insights from the cardiovascular disease in Norway project. Hypertension 76, 1506–1513 (2020).
Brumpton, B. M. et al. The HUNT study: a population-based cohort for genetic research. Cell Genom. 2, 100193 (2022).
Åsvold, B. O. et al. Cohort profile update: the HUNT study, Norway. Int. J. Epidemiol. 52, e80–e91 (2023).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Facco, F. L. et al. Association between sleep-disordered breathing and hypertensive disorders of pregnancy and gestational diabetes Mellitus. Obstet. Gynecol. 129, 31–41 (2017).
Guerrero, R. F. et al. Genetic polymorphisms associated with adverse pregnancy outcomes in nulliparas. Preprint at medRxiv https://doi.org/10.1101/2022.02.28.22271641 (2020).
Denny, J. C. et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26, 1205–1210 (2010).
Carroll, R. J., Bastarache, L. & Denny, J. C. R. PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics 30, 2375–2376 (2014).
Acknowledgements
This work was supported by grants from the US National Heart Lung and Blood Institute (K08HL166687 to M.C.H., K08HL146963 to K.J.G., R01 HL163234 to R.S. and K.J.G., R01HL139865 to R.D., R01HL155915 to R.D., DP2HL152423 to R.M.G., U01HL166060 to R.M.G., R03HL148483 to R.M.G., R01HL142711 to P.N., R01HL127564 to P.N., R01HL148050 to P.N., R01HL151283 to P.N., R01HL148565 to P.N., R01HL135242 to P.N. and R01HL151152 to P.N.); the American Heart Association (940166 to M.C.H. and 979465 to M.C.H.); the Korea Health Industry Development Institute (HI19C1330 to S.M.J.C.); Harvard Catalyst Medical Research Investigator Training Program (to A.P.P.); National Human Genome Research Institute (U01HG011719 to A.P.P. and P.N.); the Belgian American Educational Foundation (to A.S.); the US National Institute of General Medical Sciences (R35GM147197 to R.F.G. and R35GM124836 to R.D.); National Institute of Diabetes and Digestive and Kidney Diseases (R01DK125782 to P.N.); National Institute of Child Health and Human Development (R01HD101246 to D.M.H.); Preeclampsia Foundation (to K.J.G. and R.S.); Fondation Leducq (TNE-18CVD04 to P.N.) and the Massachusetts General Hospital Paul and Phyllis Fireman Endowed Chair in Vascular Medicine (to P.N.). We thank the participants and investigators from the InterPregGen consortium, FinnGen, Estonian Biobank, Genes & Health, Michigan Genomics Initiative, Mass General Brigham Biobank, BioBank Japan, BioMe, HUNT, PMBB, UK Biobank and nuMoM2b; additional acknowledgements appear in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
M.C.H., B.T. and P.N conceived these analyses. M.C.H., B.T., R.R.K., B.X., L.B., H.M.T.V., M.S.S., D.A.v.H. and T.L. performed formal analyses. M.C.H., B.T., A.P.P., R.F.G., S.M.J.C., S.M.U., K.J.G., B.M.B., S.P., S.Z., G.N.N., R.D., D.M.H., T.L. and P.N. provided resources. M.C.H., B.T., B.X., S.K., M.T., M.C.A., D.A.v.H. and T.L. performed data curation. M.C.H. and B.T. drafted the manuscript. M.C.H., B.T., R.R.K., B.X., L.B., A.S., S.K.V. and R.M.G. performed data visualization. K.J.G., R.S., G.N.N., R.D., Q.Y., I.P., S.S.V., H.C.M., D.A.v.H., T.L. and P.N. supervised the study. All authors contributed to the critical review and revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.C.H. reports consulting fees from CRISPR Therapeutics, advisory board service for Miga Health, and grant support from Genentech, all unrelated to this work. K.J.G. has served as a consultant for BillionToOne, Aetion and Roche for projects unrelated to this work. R.S. is a cofounder of Magnet Biomedicine, unrelated to this work. R.D. reports receiving grants from AstraZeneca and grants and nonfinancial support from Goldfinch Bio, being a scientific cofounder, consultant and equity holder for Pensieve Health (pending) and being a consultant for Variant Bio, all unrelated to this work. P.N. reports grant support from Amgen, Apple, AstraZeneca, Boston Scientific and Novartis; spousal employment and equity at Vertex; consulting income from Apple, AstraZeneca, Novartis, Genentech/Roche, Blackstone Life Sciences, Foresite Labs and TenSixteen Bio and is a scientific advisor board member and shareholder of TenSixteen Bio and geneXwell, all unrelated to this work. All remaining authors report no competing interests.
Peer review
Peer review information
Nature Medicine thanks Lucy Chappell, Tu’uhevaha Kaituu-Lino and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling editor: Anna Maria Ranzoni, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1
Flow chart summarizing the study design and contributing cohorts.
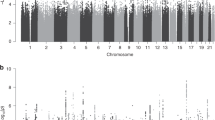
Extended Data Fig. 2 Manhattan plots of preeclampsia/eclampsia and gestational hypertension in discovery cohorts.
Manhattan plots (chromosomal position on the X-axis and -log(10) of the P value on the Y-axis) are displayed for (a) preeclampsia/eclampsia in 17,150 cases and 451,241 controls and (b) gestational hypertension in 8,961 cases and 184,925 controls. Analyses included multi-ancestry meta-analysis of common variants (minor allele frequency ≥1%). Loci are labeled by the gene nearest to the lead variant. Two-sided P values (not adjusted for multiple comparisons) are from Z scores from fixed-effect inverse-variance weighted meta-analysis.
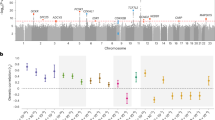
Extended Data Fig. 3 Results of multi-trait analysis of genome-wide summary statistics (MTAG) for preeclampsia/eclampsia.
Results are from joint analysis of summary statistics for preeclampsia/eclampsia and gestational hypertension in discovery cohorts. The plot displays chromosomal position on the X-axis and -log(10) of the P value on the Y-axis. Two-sided P values (not adjusted for multiple comparisons) are from Z scores from MTAG.
Extended Data Fig. 4 Relative expression of prioritized genes in human aortic cells with single-nuclei RNA sequencing.
We analyzed expression of genes prioritized by genome-wide meta-analysis of preeclampsia/eclampsia and gestational hypertension and secondary in silico analyses in a dataset of single-nuclei RNA sequencing from two normal human flash-frozen aortic specimens. Most prioritized genes were enriched in endothelial cell populations and/or macrophages.
Extended Data Fig. 5 Sex-stratified phenome-wide association study of gestational hypertension polygenic risk in the UK Biobank.
Gestational hypertension polygenic risk was associated with 1,445 phenotypes among (a) female and (b) male participants in the UK Biobank. Associations with phenotypes were tested using logistic regression with adjustment for age and the first five principal components of genetic ancestry. Two-sided P values (not adjusted for multiple comparisons) are from logistic regression models adjusted for age and the first five principal components of genetic ancestry.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Note.
Supplementary Tables
Supplementary Tables 1–19.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Honigberg, M.C., Truong, B., Khan, R.R. et al. Polygenic prediction of preeclampsia and gestational hypertension. Nat Med 29, 1540–1549 (2023). https://doi.org/10.1038/s41591-023-02374-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-023-02374-9
This article is cited by
-
Parental genetically predicted liability for coronary heart disease and risk of adverse pregnancy outcomes: a cohort study
BMC Medicine (2024)
-
Sex-specific genetic architecture of blood pressure
Nature Medicine (2024)
-
Using the methylome to predict pre-eclampsia
Nature Medicine (2023)