Abstract
Cropping system models are deployed as valuable tools for informing agronomic decisions and advancing research. To meet this demand, early career scientists are increasingly tasked with building crop models to fit into these system modelling frameworks. Most, however, receive little to no guidance as to how to do this well. This paper is an introduction to building a crop model with a focus on how to avoid pitfalls, minimize uncertainty, and maximize value. We synthesized knowledge from experienced model builders and literature on various approaches to model building. We describe (1) what to look for in a model-building dataset, (2) how to overcome gaps in the dataset, (3) different approaches to fitting and testing the model, and (4) how to avoid common mistakes such as over-parameterization and over-fitting the model. The process behind building a crop model can be overwhelming, especially for a beginner, and so we propose a three-pronged approach: conceptualize the model, simplify the process, and fit the model for a purpose. We revisit these three macrothemes throughout the paper to instil the new model builder with the methodical mindset needed to maximize the performance and impact of their crop model.
Similar content being viewed by others
1 Introduction
Crop models have the potential to transcend our understanding of how crops interact with agronomic management across space and time. Models have been accepted as useful tools that help agronomists, farmers, policy makers, and other researchers make more informed decisions and recommendations. In this paper, we provide a guide on how to build a process-based crop model within a larger cropping system framework. There are many different kinds of crop models, and so, we want to be explicit in defining the scope of this paper: this paper is not necessarily applicable to 3D-crop modelling or machine learning, although many of the basic principles around quality control and model testing are universal for model building.
Building this type of crop model requires finding a balance between theory and practicality, complexity and certainty, breadth, and depth, such that the model can capture both the internal plant mechanisms driving growth and the external interactions of these mechanisms with a dynamic cropping system. In their 2012 review, Prost et al. called for more debate and discussion about the model design process. This paper’s aim is to invite more early career model builders into that conversation, giving an overview of the model building landscape: what to look for in a dataset, what questions to ask, and how to test if your model is good.
Many of the examples referenced in this paper are sourced from the Agricultural Production Systems sIMulator (APSIM; Holzworth et al. 2014; 2018); however, the principles are applicable to building a crop model within other cropping system frameworks (e.g., STICS (Brisson et al. 2003) and DSSAT (Jones et al. 2003)).
The goal of a crop model is to simulate plant growth as the product of a series of interactions among the plant, soil, climate, and management (Hornberger and Spear 1981). The model itself is a complex web of simple algorithms, each describing a different interaction and tying all the model components together. Within the plant itself, the allocation and reallocation pathways of biomass and nutrients are a constantly changing and interlocking network of supplies and demands. Because of this tight coupling, inaccuracies or uncertainties in one process will be potentially compounded in another process downstream. Each step in the process of model building should be taken with the aim of making the model as simple and data-driven as possible to reduce the risk of such uncertainties (Stirling 1999). Moreover, given the interwoven nature of the different components within the model, diagnosing the source of these uncertainties becomes increasingly difficult as the model grows in complexity (Balci 1995).
“Garbage in, garbage out” is a common saying among model builders; i.e., a model can only be as good as the data you use to build it. We propose that the best approach to building a model should extend beyond the data itself. This approach is particularly useful when modelling with a sparse dataset as would be the case with a crop that has not been extensively researched. The dataset you use, much like the code the model is comprised of, defines the boundaries and limitations of the model (Boote et al. 1996; Brisson et al. 2003). Meanwhile, the quality of a model is akin to how useful and robust it is.
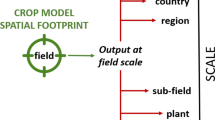
The process of building a model should start before the model-building dataset is compiled with a concept of how the model should work. You, the model builder, should then use the data to focus that concept, break the model down into simple parts, and fit each part for a purpose, be it minimizing uncertainty within the model or targeting the needs of the end users. The aim of this three-pronged approach (conceptualize, simplify, and fit for purpose) is to avoid over-parameterizing the model and to maximize its impact (Fig. 1). We will revisit this approach throughout this paper as we delve into the specifics of building a crop model.
2 Getting started
2.1 Conceptualize: what are the needs of the potential model users?
There are many potential uses for a crop model, but most model builders aim to build a model that is versatile enough to both improve understanding and be a tool for action; achieving both aims, however, requires simultaneously targeting two user groups with different needs who operate often at different scopes (Boote et al 1996; Prost et al. 2012).
As a model builder, you want to gain the confidence of those who will use your model. To gain this confidence, you must understand what they need the model to simulate and construct a model structure that can fill those needs. While this may seem obvious, Prost et al. (2012) found that only 30 models out of the 518 they reviewed included input from the targeted users in the design of the model. It is important to determine what crop responses to model to meet the needs of the user and what data is necessary to inform those responses (Boote et al. 1996). For instance, grain N concentration response to management is quite important to a chickpea researcher focused on human nutrition, but perhaps not so important to a maize researcher focused on ethanol. For this purpose, consider what the scope of the model needs to be, i.e., how detailed or general the model needs to be. Increasing the level of detail in the model increases the risk of error (uncertainty) and reduces the model’s stability (versatility) when applied in a more general context (Saltelli 2019). Once you understand the model’s purpose and scope, you can think about what data you need to fit that purpose and how data availability may limit the scope.
2.2 Simplify: aligning the needs of the model with the availability of data
2.2.1 Minimize uncertainty
When laying the foundation for your model, you want to maximize versatility and minimize uncertainty (Fig. 2). Increasing model complexity can increase the versatility or scope of the model (how many questions the model can answer) and decrease the sensitivity of the model to individual inputs, but it also increases the model’s overall uncertainty (Muller et al. 2011). The model’s overall uncertainty is the cumulative uncertainty of its processes (phenology, canopy development, etc.) (Snowling and Kramer 2001). The baseline experiment you use to fit each process within the model should be aimed at minimizing this uncertainty by being akin to a control plot in a field experiment. Think optimal, well-documented conditions (well-watered, fertilized), a single crop variety (preferably one you know a lot about), no extremes in terms of management (e.g., plant population and row spacing), and no biotic stresses (e.g., pests and disease). Removing stress from the environment removes uncertainty: stressed treatments require you to simulate not only the plant response to stress but also the stress drivers correctly. Like a control plot in a field trial, building the foundation of a model with data from a simple predictable experiment (or, at least, an experiment where there are fewer variables at play) provides a good baseline for understanding how the plant develops (van Ittersum et al. 2003). Additional complexity (e.g., response to nutrients or sowing time) should be added incrementally, each time with a focus on minimizing uncertainty (Snowling and Kramer 2001).
2.2.2 High-quality inputs
Any linguist, anthropologist, or politician will tell you: context is important. In a model, the context is wherever the plant is growing in the experiment you are simulating. The corresponding environmental inputs pertaining to initial soil resources, climate, and management feed into the model. The algorithms and parameters that make up the model dictate how the plant responds to the inputs. Inevitably, there will be errors in the inputs, but some in the crop modelling community are starting to develop standards for metadata (https://dssat.net/data/standards_v2/) in an attempt to alleviate these errors. In general, try to minimize these errors via careful scrutiny of your data before fitting your model to it: assess the level of uncertainty associated with your data sources or the resolution of the data and look for human error in both the collection and simulation of the data (Ojeda et al. 2021).
Soil-plant interactions in the model are dictated by the balance between the supply and demand of key resources (e.g., nutrients, water) (Brown et al. 2009, 2019; Monteith 1986). Detailed soil data, however, is often difficult to find. Simulating a dataset that includes initial soil nutrient and soil water measurements is ideal. When this data is not available, estimates of these inputs can be made using a deductive approach we will discuss later in the section on simulation-specific parameters.
Most crop models require daily rainfall, temperature (maximum and minimum), and solar radiation data. As these data inputs are site-specific, any discrepancy between where they were measured and the corresponding plant and soil data collected should be noted as a potential source of error. Rainfall (and irrigation) brings more water into the system; temperature drives how fast the nutrient pools turn over and the plant develops; radiation drives evaporative demand and plant photosynthesis. If the climate data is incorrect, the model will miscalculate how quickly the plant develops and what resources are available to it. Spend some time checking the climate data you have, looking for anomalies or extremes, and perhaps compare it to another reference set (e.g., another dataset from nearby) if you have one.
For every experimental dataset you access, there is a variety of metadata (i.e., details about the experimental management, environment, and data collection methodology) that you will invariably require in your model building. Either in the paper or dissertation itself or through correspondence with the data custodian, you need to find out the dates and details of all management activities in the experiment. This can include, but is not limited to, crop variety name, sowing date and depth, row spacing, plant population, fertilizer and irrigation amount, type, and application date(s), in-season sampling dates, tillage, and the previous crop history. Should you not be able to acquire this additional data, consider how sensitive the simulations are to any input data that need to be estimated or deduced. If the model is overly sensitive to the uncertainty surrounding these estimated inputs, reduce how much you rely on that experiment in your model calibration. Such data with higher levels of uncertainty, however, may still be useful for testing the sensitivity of your model.
2.2.3 Focus on the processes
Process-based crop models need time-series data points throughout the season to describe/represent/simulate plant development and growth. As few biological processes are linear, a series of data points (often at least five to six) taken at different times can map out the correct curvature of a relationship describing plant development.
There needs to be a balance between the level of detail in your dataset and the level of complexity in your model to minimize model uncertainty (Brisson et al. 2003). In modelling plant development, consider what processes are key for the model user and the corresponding availability of relevant in-season measurements (e.g., canopy development, biomass, and nutrient allocation/reallocation). The more complex the model, the more quality data is required for the model to maintain enough certainty to answer relevant questions precisely and accurately (Saltelli 2019; Gaetani et al 2020; Muller et al 2011). No matter the process, look for data that was collected multiple times during the season and experiments that measured more than one attribute at the same time; such data will help you dissect the mechanisms and pools behind the processes.
2.2.4 Quality control
No dataset is perfect, but the impact of whatever errors exist in it should be monitored. One monitoring approach is outputting biomass pools or water balance over time to see if the numbers and growth/uptake rates seem reasonable (i.e., in accordance with what has been recorded in the literature for the targeted or similar crops) or to confirm that resource supply and demand relationships (both among the plant organs and between the plant and soil) are balanced. Visualizing the data is a particularly effective way of locating outliers and errors and is an approach which should be used routinely throughout the model-building process.
If something in the dataset seems wrong, it is ideal if you can ask the experimentalist about it. If this is not possible, find the source of the error by focusing on the main drivers of variability in the data. For instance, if the experiment is rainfed, rainfall will be the largest water balance term, not drainage or evaporation. In this case, look at the rainfall data before looking at the soil profile or radiation data. If that experiment were to be irrigated, the timing/amount of irrigation may be the source of the error, not the rainfall data. Simple conceptual models can be used to check that the resource allocation within the system is balanced e.g. that the crop is not taking up more water than is available, etc.
2.3 Fit for purpose: work within the dataset’s limitations
No matter how much data you aggregate for building your model, there will be gaps of knowledge in your model-building dataset (Boote et al. 1996; Snowling and Kramer 2001). A model-building dataset, however, should include a variety of experiments that, together, cover both the most relevant measurements and a diversity of environments and management combinations. In experiments with wide breadth (multiple locations, varieties, treatments, etc.), the field researcher likely did not take many in-depth measurements. In contrast, highly detailed studies measuring a range of critical plant and/or soil processes are unlikely to be repeated across multiple environments. Find a balance between the two: focus on the process the data describes rather than the data itself, and understand the limitations of your dataset. A good dataset includes enough specifics about the corresponding environments, experiments, and plant development to allow you to decipher trends and relationships but is broad enough to give you confidence that those trends and relationships are consistent, not outliers.
At the end of the day, it is important to find value in the data you have while recognizing its shortcomings. Employing a sliding scale of how much trust you put into a given dataset allows you to decide how much weight you put on it for model development. Identify your ‘anchor’ datasets i.e. relevant datasets you are familiar with and have a lot of faith in their quality. These are the datasets you should care the most about the model capturing. Together with the model structure, the quality of these datasets is important for minimizing model uncertainty. As the trust or relevance in a dataset declines, you, as the model builder, can become more comfortable in accepting poor fits and moving on. This process is subjective, but it is important to not get caught up in extraneous detail. Remember, future experiments can target any holes in this initial dataset. For now, start simple and build with purpose.
3 Fitting the model to the data
3.1 Conceptualize: how should the model run?
3.1.1 Start with an existing crop model
Start building by using the framework of an existing crop model, preferably a crop similar to the one you are trying to simulate (Holzworth et al. 2018). There are common processes that span across multiple crop species and thus across different crop models (Brown et al. 2018; Robertson et al. 2002a; Wang et al. 2002). Building on the progress made by other model builders also helps you stay on the forefront of modelling, increases your confidence that the software works and makes it easier to get your finished model published as you can cite the foundational work. This approach is applicable even if you are just building or adapting a section of a current crop model: look at the relevant sections in preexisting models of similar crops.
In the process of fitting the original model to your data and crop, you will likely change multiple aspects of the model. With each change, however, be methodical and try to retain any value the old model has to offer; use version control to make it easy to revert to earlier versions as needed. As no dataset is comprehensive, look for where other crop models may have taken an approach or defined a parameter value that could fill-in a deficiency in the model-building dataset you compiled (Bouman et al. 1996).
3.1.2 Equifinality
Especially when faced with modelling data from more complicated experiments, it is tempting to fit the model to the data irrespective of logic or reasoning. To guard against this temptation, throughout the model-building process, revisit your initial simple experiment to perform sensitivity tests. Take the simple template and adjust the conditions (water, temperature, plant population, etc.) one at a time, making sure the model responds logically. In process-based models, such sensitivity tests can ensure the process, not just the results, are correct or at least reasonable.
Many modellers rely on an educated guess-and-check approach when calibrating a model’s parameters for a given experiment (Seidel et al. 2018). While not ideal, this approach is likely necessary when it comes to parameters that are more abstract (e.g., photo-thermal-vernal days to floral initiation). If the parameters relate to something that is easily measured and conceptualized (e.g., final leaf number under short and long days with and without vernalization), they should be able to be deduced easily from data or expert knowledge. Parameterize your model with data at each stage as much as possible, building upon the foundation of the previous stage.
Equifinality is the idea that in an open system, there is more than one potential way to reach a desired end result (Beven and Freer 2001). As a model builder, if you focus on the end result (e.g., yield) and not the processes leading up to it, you may end up with a model that can simulate yield accurately in one location/season, but has little value outside that setting. Meanwhile, if you fit the model to capture both leaf area index (LAI) throughout the season and yield, the potential range of acceptable parameter combinations declines. In this sense, having a diverse dataset is crucial for decreasing model ambiguity. If you get the process right, rather than just trying to hit the target any way you can, your model will be sound and robust.
3.2 Simplify: build chronologically and focus on one process at a time
3.2.1 Appropriate parameterization
To fit a model to data, it is necessary to understand which parameters are specific to a simulation and which are part of the overall crop model (i.e., consistent across simulations) to avoid over-parameterizing the model. The structure of the model defines how processes in the model interact with each other and site-specific parameters and inputs. Depending on the cropping systems framework you are building for, your model’s structure might be driven by a single primary process like deWit’s model, wherein all processes and parameters build off of photosynthesis (Bouman et al. 1996) or be more adaptable depending on the complexity of the crop or available dataset like the STICS model (Brisson et al. 2003).
Simulation-specific parameters: characterizing your soil
Before you fit the overall crop model parameters, make sure that you are confident in your simulation-specific parameters. In APSIM, these parameters include soil upper and lower water limits, soil hydraulic conductivity, C:N ratio, and initial organic matter and N levels for each layer of the soil profile (the number and thickness of each layer is user-defined). The degree of detail needed for these parameters depends on the modelling framework you are building for.
In general, how well a plant grows is often dependent on how much nutrients and water can be sourced from the soil. You need to know more than what these soil components (nutrients and water) were at one point in time: you need to know the dynamics of how those components change and interact with each other over time. Setting these soil parameters may prove to be difficult as most of the field studies populating your model-building dataset likely do not have detailed soil nutrient and water data. Moreover, where some soil parameters can be measured directly (e.g., soil upper and lower limits), others are more conceptual (e.g., soil C and N pool decay over time), requiring inferences and deductions be made, drawing from theory and data from similar environments to supplement whatever data you have for your study.
Just as we recommended building your model off an existing crop model, look for an existing soil (perhaps from a public soil database or a simulation built for another model) to begin the process of parameterizing your simulation site’s soil. Use this soil to run a simulation in a tested crop model of a common crop like wheat to check if the model outputs logical values before using that soil as a foundation for building your crop model. As you did with the inputs, use a visualization approach to check for errors and missteps made in the copying and modifying of any configuration from another simulation (Ojeda et al. 2021).
Depending on what data is available to you, the approach you take to customize the foundational soil may vary. For example, when parameterizing the soil C and N pools, Kivi et al. (2022) used data from multiple sites: they took initial soil C and N and in-season soil water measurements from their targeted site, but then supplemented that data with information from similar well-documented sites. Moeller et al. (2007) used data from multiple treatments: they found that plant N uptake data in irrigated treatments where no N fertilizer was applied could be used to parameterize the rate of N mineralization. Huth et al. (2010) used multiple years of soil C and N data to calculate the rate of soil C and N decay and, thus, deduce how much of the C pool is inert vs. labile and the soil C:N ratio over time. All three studies applied a conceptual understanding of how C and N flow through the system to parameterize the soil dynamics.
There are also many valid approaches to deducing soil water parameters when available data is limited. Angus et al. (1980) and Peake et al. (2010) demonstrated the merit of deducing your soil’s parameters from the soil parameters for other sites with similar soils. Hochman et al. (2007) parameterized plant available water in soils with subsoil constraints by applying theoretically derived modifiers to default values extrapolated from top soil measurements. Taking a more comparative approach, Whish et al. (2014) parameterized soil water uptake by looking at soil water data across paddocks with different levels of nematode infestations, where conceptually, they knew that a larger infestation would lead to less water uptake. Similarly, Monteith (1986) and Brown et al. (2009) examined the balance between water supply and demand across different water regimes. Looking at water dynamics on a larger scale, Zhang et al. (2001) used data from 250 grassed catchments and theory from previous studies to parameterize a model that relates rainfall and evapotranspiration.
Part of the deduction approach is applying periodic logic checks throughout the parameterization process to make sure a given parameter value makes sense in the system. Such logic checks on crop-soil-climate interactions should play an active role in any model development, including crop models outside of cropping system frameworks. For example, when parameterizing soil N pool fractions and turnover rates, check if the values associated with all components contributing to the N balance (e.g., microbial N and leaching) are sensible. If not, determine what components need to be adjusted and how are they influencing the others. Change the parameterization to reflect which factor logically dominates the soil N dynamics. Run such logic checks on the parameters throughout the building process as changing something in one part of the model may impact the dynamics of another part.
Model parameters
Once you are confident in the inputs and simulation-specific parameters, you can move onto the model parameters (parameters that make up the crop model itself, not specific to any one location or experiment, e.g., phenology, rate, and pathways of plant development). Model parameters dictate how crops respond to the environment and management they encounter.
As the output from one process may impact the inputs to another, start with the most independent processes before moving onto the more dependent processes. For example, phenology is a largely independent process as it is rarely impacted by plant growth and resource availability. On the other hand, plant growth is a more dependent process as it is driven by phenology. Likewise, as mentioned earlier, start the building process by parameterizing potential growth and yield-forming processes for stress-free treatments. To model stress responses is to model a deviation of the processes away from their potential. If you do not have the potential correct, it is impossible to correctly parameterize stress responses in your model. In this section, we will discuss the processes in the order we suggest you parameterize them.
3.2.2 Phenology
There is often discrepancy between the resolution at which the model predicts phenology and the data that is available. Should your model predict the timing of emergence, end juvenile, floral initiation and flowering, but your model-building dataset only has data for flowering timing, there are a lot of parameter combinations that will fit the data well (He et al. 2017). To minimize the uncertainty around phenology, begin with parameterizing phenology chronologically from sowing to maturity.
The primary metric of development is cell division, driven by temperature (often defined as thermal time). The first step in defining phenology is determining what time interval you will use to calculate thermal time. Within a given cropping systems framework, the time interval should be consistent across all crop models. From the literature (or your own experiments), determine the base, optimal, and maximum temperatures for plant development (Ritchie and NeSmith 1991; Robertson et al. 2002b; Yin et al. 1995). Use the recorded stage dates to build a thermal timeline (i.e., the accumulation of thermal increments over the growing season).
For example, APSIM calculates thermal time in 3-hour increments as an interpolation of the minimum and maximum air temperatures (Jones et al. 1986). The rate at which thermal time is accumulated is the response variable of a simple stepwise linear regression that relates the aforementioned base, optimal, and maximum temperatures to the thermal time. The rate of accumulation as well as the threshold temperatures may change during the life-cycle of the plant.
In general, starting from sowing, determine how long it takes the plant to get from point A to point B on that timeline. Consistency in scoring and staging the plant is important: if you are, or can consult with, the experimentalist, developing a detailed guide for the people recording the phenology will increase the confidence you have in the data. That said, in situations where the phenology data was sourced from previous studies, consider the level of ambiguity in some of the phenological data (e.g., the definition of 50% flowering date) and steer toward quantitative data (e.g., number of main stem nodes) as much as possible. The high level of uncertainty surrounding phenological parameters, often due to the strong influence of genotype by management by environment interactions and discrepancies in measurement methodology, may be reduced after years of additional model-testing field experiments as was the case with the APSIM canola model (Robertson and Lilley 2016) or may require statistical or machine learning calibration approaches such as those demonstrated with APSIM Oryza model (Nissanka et al. 2015) and with the APSIM Maize model (Akhavizadegan et al. 2021). It is important to remember that model construction is an ongoing process (Boote et al. 1996).
While thermal time controls the speed of movement toward a target on the phenological timeline, vernalization requirements and photoperiod sensitivity can alter the size of the target. Create a conceptual model capturing the factors you expect to influence phenology. Use data from a nonstressed treatment to define the degree to which the plant is sensitive to these factors; it may vary depending on the variety, location, and year.
A threshold amount of cumulative thermal time units is needed for any plant to begin its reproductive stage, but for some plants, this amount lessens with the accumulation of thermal units within a range of colder temperatures in a process known as vernalization. Similarly, the duration of the daylight hours can impact the rate at which thermal time units are accumulated if the plant is photoperiod sensitive. For vernalization and photoperiod sensitivity, modifiers can be applied to adjust the baseline phenology relative to the temperature or photoperiod, respectively. Initially, it is best to be conservative with these modifiers: it is rare that the plant in the field is responding to just one factor at a time (Zhao et al. 2014). For example, while it may appear that colder temperatures are accelerating phenology, this impact may be exacerbated by some stress (pertaining to a disease, pest, water, etc.) that exists in that one location but not in another.
3.2.3 Canopy development and biomass accumulation
After defining the thermal time targets for each phenological stage, it is time to outline how the plant develops during those time intervals. Map out conceptually how you expect the plant to develop and fit the parameters in a chronological order as early development will impact later development.
Within each developmental stage, simplify the process of parameterization by focusing on one morphological process at a time, starting with the most independent: canopy development. As canopy development is directly related to phenology, parameterize the leaf, tiller, and/or branch appearance rate(s) relative to the nodal appearance rate. If you do not have that level of detail in your dataset, take a simpler approach and focus on full canopy development using LAI and/or radiation interception data.
In APSIM’s canola model, phyllochron and LAI could be measured easily, but leaf area varied depending on the location of the leaf on the plant and the carbon supply (Robertson and Lilley 2016). As such, a reasonable maximum value for the specific leaf area (SLA) was estimated from existing literature and models of similar crops. The model builders then deduced the rate and extent to which this maximum value needed to decrease as carbon supply decreased and the canopy filled such that the model simulation reflected the measured LAI and SLA data collected at different plant densities.
After canopy development, focus can shift to plant growth and the allocation of biomass within the plant. A radiation use efficiency (RUE) value can be sourced from a field study, but the idealized RUE value that the model will use to calculate potential plant growth is likely higher than what was measured in the field. Once you have total biomass accumulation from the RUE, consider the allocation of that biomass to various organs within the plant. Ask the question: at different stages of development, what fraction of the total biomass is allocated to the stem vs. the leaves, reproductive components, etc.?
In general, while preflowering development occurs independently of growth, the rate of development and growth interact strongly post-flowering (Brukhin and Morozova 2011). When transitioning your focus from thermal time-driven development to plant growth, consider the role the resource availability of radiation and water play in these processes.
Ideally, within each plant organ, you want to know how much of the biomass can be reallocated to other organs (termed in APSIM as the metabolic and storage pools) to meet supply/demand requirements and how much cannot (structural pool, biomass necessary for plant growth). In general, however, there is a lack of data about the mechanisms driving biomass allocation. As with defining the soil parameters, there are multiple ways to determine the relative size of these pools. If you have partitioned biomass measurements from multiple points during the season, graphing the relationship between the organ biomasses or an organ biomass against the total biomass can provide insight to those relationships at different points of time. Alternatively, or additionally, if you have organ nutrient concentrations over the season, you can infer the fraction of biomass allocation or reallocation needed to result in the changes in concentration over time.
In order to determine the source and sink dynamics of N remobilization during the grain filling in APSIM-Nwheat, Asseng et al. (2002) derived values for how assimilates were remobilized from measured differences in the water soluble carbohydrates and protein carbohydrates between anthesis and maturity in the stems, spikes, and leaves. To reduce the risk of overfitting the model to one experiment/environment, they repeated these measurements in two cultivars at three different N fertilizer rates in three field locations. This approach to modelling N remobilization in APSIM-Nwheat was adopted and modified from a preexisting model (CERES Wheat, Jones et al. 1983) and a detailed review of existing literature on N remobilization in wheat across different environments informed what field measurements could be used to calibrate the model. Asseng et al. (2002) then ran multiyear simulations in sites where field data had not been collected to see if the simulations agreed logically with what had been reported in the literature. From these simulations, they could better understand the extents of the model’s versatility and bias.
Without such detailed data, some crop modellers make assumptions about the functional balance needed between organs to optimize crop growth while others look for similarities between their crop and others to surmise generalized partitioning routines (Brown et al. 2019). Irrespective of the method, run simple logic checks within your crop model to make sure that conceptually the biomass is not being overall located to any organ.
3.2.4 Senescence and stress
Applying senescence should be the final step of building the baseline model. As quality data on the amount of senesced biomass is difficult to obtain and frequently lacking, a deductive or simplified approach may be necessary to parameterize this process. If some of this data is available, you can derive the timing and rate of senescence from the balance of the remaining green/active biomass. Without such data, the simplest approach to senescence is setting it as a linear progression from some initiating stage to when the crop can be harvested.
After completing the parameterization of the baseline model with data from nonstressed experiments, you can start fitting the model to data from stressed experiments. First, set simulation-specific parameters to make sure you are capturing the stress accurately. Then, you can apply modifiers to delay phenology, reduce biomass accumulation, or accelerate senescence in response to a given stress. These modifiers can be stage-dependent: at some stages of development, the plant may be particularly vulnerable to stress (e.g., setting grain number) and at others it may be able to recover. If plant response to stress is dependent on the level or timing of the stress, you can define a stress level indicator to feed into stage-dependent linear interpolations of plant response to the stress. If the plant response is more definitive, set limits of threshold values above or below which the plant dies or growth ceases.
3.3 Fit for purpose: sensitivity and uncertainty analyses
No model is perfect. It is valuable to know the parametric source(s) of uncertainty in the model (Muller et al. 2011). A sensitivity analysis looks at that uncertainty, detailing how much the output of the model changes as the result of changing one or more parameters (Beven 2018; Monod et al. 2006; Muleta and Nichlow 2005; Saltelli 2002). Ranking the importance of the parameters based on how sensitive the outputs are to changes in the parameters is a common way of conducting a sensitivity analysis (Monod et al. 2006; Muleta and Nichlow 2005; Saltelli 2002). Two common versions of this approach are the Morris and Sobol methods (Morris 1991; Sobol 2001). The Morris method is a one-at-a-time approach that looks at the relative response of the model outputs to a change in a single parameter. The Sobol method looks at how much the model output responds to variability in a single parameter vs. to the interaction effect that variability has with the other parameters. Repeated random sampling approaches like Monte Carlo simulation or parametric bootstrapping are valuable as they increase the probability that the parameter sensitivity is consistent across a range of contexts.
4 Testing your model
Any publication or presentation of a new crop model will make that model look as good as possible. To make sure that your model is actually good (and to determine how good another model really is), it is important to apply critical thinking to what tests are used to evaluate the model’s performance and what the test results mean (Gauch et al. 2003).
The aim of testing your model is to see if the parameters calibrated for the model-building dataset are relevant for data outside of that dataset. This is done by evaluating how well the model can simulate data outside of your model-building dataset, preferably data that comes from a range of environments and is relevant to the targeted model users’ systems (Gauch et al. 2003). There are multiple statistical approaches to testing a model, but none of them are comprehensive: each test will convey a different aspect of the model’s accuracy or lack thereof. It is also important to contextualize the resulting graphs or indices as some approaches are sensitive to outliers and others to the size of the dataset. As there are strengths and weaknesses to every testing approach, it is best to use more than one (Wallach 2006). Applying more than one test can shed light on the source of the error, the residual error (i.e., how close the simulated data is to the real data), and whether there is a bias in the model (i.e., whether the model outputs trends higher or lower than the real-world data).
4.1 Conceptualize: what are you testing?
There are two primary methods for selecting what data will go into your testing dataset: the split dataset and the independent dataset. In a split dataset, the data used to build and test the model source from the same studies. The subset of data that is used for testing should be selected at random (e.g., bootstrapping) (Liu et al. 2018; Wallach 2006). Alternatively, you can test the model with a dataset that is completely independent from the initial model-building dataset. Both approaches are valid, although the latter may demonstrate the scope of the model better because it would bring more variability in the environment, management, and climate into the equation with data from a new set of independently conducted field experiments.
4.2 Simplify: graphical and statistical analyses
There are many publications that go into detail about what test is best to use for what dataset/model (Gauch et al. 2003; Monod et al. 2006; Moriasi et al. 2007, 2015; Wallach 2006; Yang et al. 2014). Here, we will briefly discuss the most commonly used/recommended tests for crop models and the reasoning for them.
Before applying statistical tests to your model, take a graphical approach to look for visual agreement between the observed and simulated data (Moriasi et al. 2015; Wallach 2006). Graphical time series visualization can pinpoint where the observed and simulated data start to disagree and, thus, reduce the risk of equifinality. For example, when evaluating simulated data across a temporal scale, graphing the data would show if the error lies in the rate of plant development or the allocation of resources at a given time.
Commonly, model builders regress simulated data against the corresponding observed data in what is called “predicted-observed graphs” with a 1:1 line plotted for reference. There is much debate about the orientation of the plot axes, i.e., whether to put the predicted or observed data on the x-axis (Piñeiro et al. 2008). Correndo et al. (2021) discussed the importance of reflecting on the unknown uncertainty level in both the predictions and observations in order to better understand the model’s accuracy and precision as independent metrics of its performance. For this, they recommended using the Standardized Major Axis (SMA) linear regression model that symmetrically takes into account error in both the predicted and observed data with respect to the 1:1 line, removing the aforementioned ambiguity of axis orientation. SMA, however, is sensitive to the size of the dataset and should not be used with smaller (n<20) datasets.
It is important to not just visualize the relationship between the predicted and observed data, but also the differences. While quantitative methods discussed later in this section analyze the average difference between the predicted and observed data, Bland Altman (1965) plot analysis, like the time series graphs, shows if there is any relationship between the model errors and where it occurs. Bland Altman analysis plots the difference between the predicted and observed against the mean to see if there is a bias toward lower or higher values and the distribution of the frequency of differences to check for normality (Bunce 2009; Giavarina 2015).
The most common statistical tests for process-based models (as well as machine-learning models) are Root Mean Squared Error (RMSE) and the coefficient of determination (r2).
RMSE (Eq. 1) is valuable for its real-world practicality as it outputs the error in the units of the measurement. When communicating with the stakeholders of the model, conveying the model error in real-world units can increase both their understanding of and confidence in the model.
Meanwhile, r2, while highly relatable given the prevalence of its usage in general and acts as more of a measure of precision than accuracy (Bunce 2009; Gauch et al. 2003; Lemyre et al. 2021; Piñeiro et al. 2008; Yang et al. 2014). This metric should be limited to use in presentations to a lay audience, rather than a singular measure of model accuracy.
A major shortcoming with both RMSE and r2 is that they are sensitive to outliers and assume that the observed data is consistent with minimal variability within itself. Alternative commonly used and recommended statistical options that analyze the model accuracy relative to the precision of the observed data are the Nash-Sutcliffe efficiency (NSE), Root Mean Squared Deviation (RMSD), and RMSE-observation standard deviation ratio (RSR) (Moriasi et al. 2007; Piñeiro et al. 2008).
NSE (Eq. 2) normalizes the residual variance between the observed and simulated data to the variance within the observed data. This statistical output shows the flexibility of the simulated data to fit the variability in the observed data, not just capture the mean. NSE, like r2, captures how closely the simulated vs. observed data fits a 1:1 line. The higher the NSE, the better, but it is important to keep in mind that the size of the dataset or natural variability of some types of measurements may produce low values irrespective of the model’s performance level (general performance ratings for NSE values are 0.75 < NSE ≤ 1.0 for Very Good; 0.65 < NSE ≤ 0.75 for Good; 0.5 < NSE ≤ 0.65 for Satisfactory; NSE ≤ 0.5 for unsatisfactory; Moriasi et al. 2007).
RMSD (Eq. 3) reports the mean deviation between the predicted values and the 1:1 line of a predicted-observed regression where the predicted values are graphed on the x-axis (Piñeiro et al. 2008). Meanwhile, RMSE relates predicted values to the predicted-observed regression line, providing an underestimation of the error between observed and predicted values.
RSR (Eq. 4) normalises RMSE to the standard deviation of values in the observed data. While the lower the RMSE, the better, a low RMSE does not necessarily mean that the model is capturing the dynamic variability of the system if the variability in the observed dataset is low (general performance ratings for RSR values are 0 ≤ RSR ≤ 0.5 for Very Good; 0.5 < RSR ≤ 0.6 for Good; 0.6 < RSR ≤ 0.7 for Satisfactory; 0.7 < RSR for unsatisfactory; Moriasi et al. 2007).
Percent bias (PBIAS) (Eq. 5) looks at whether the simulated data tends to trend higher or lower than the observed data, where a value of zero means a perfectly accurate model, positive values mean the model is underestimating the observed data, and negative values mean that it is overestimating. PBIAS is particularly useful in ensuring that the supply/demand dynamics are balanced in the model. While the closer to zero the PBIAS is, the better, the threshold values for the performance ratings are dependent on what is being measured (Moriasi et al. 2007). In crop models, we expect the model to be biased, however, and generally to overestimate yield, biomass and leaf area since few models (at the time of this publication) account for the biological limitations of pests and diseases. In the future, this bias may lessen, but it will likely always exist as few biological pressures are easily and consistently detected in the field.
4.3 Fit for purpose: scenario analysis
You may not have data to test all the conditions relevant/important for the targeted model users. It is still important to make sure that the model works well in those conditions and to test the limits of the model (Holzworth et al. 2011). Setting up scenario analyses and confirming that the model’s outputs are logical will increase the users’ confidence in the capacity of the model to perform well outside of its building and testing datasets (Ojeda et al. 2021). Data trends relative to other similar (or known) crops can also help confirm the logicality of these theoretical tests. For instance, simulate crop growth in an environment where the crop should not be able to grow well or at all: if a model were to let a summer crop grow well in the winter, there is likely something wrong with that model.
5 Conclusion
Building a crop model is a difficult task and one that will never be completed. As more data becomes available, the more the model can be improved, as long as the foundation is strong. Just as there is no perfect model (or experimental dataset), there is no one perfect approach to building a model. The aim of this paper was to provide a basic roadmap for a model-building journey by suggesting what kind of questions the model builder should ask, how to work around data gaps, and how to test the validity of the model.
We propose a three-pronged approach to model building: (1) conceptualize the model before you start to build so you know what data you need to achieve the desired aim and scope of the model, (2) simplify the model-building process by focusing on one process at a time to guard against the pitfalls of over-parameterization and equifinality, and (3) fit for purpose so that the model is targeted and meets the needs of the users. The foundation of this approach is based on the belief that the process of building a model needs to extend beyond fitting the model to a dataset. Each step in the building process should be taken methodically with a focus on both data and logic rather than expediency. Our approach aims to minimize uncertainty and maximize utility in the model.
Data availability
Not applicable.
Code availability
Not applicable.
References
Akhavizadegan F, Ansarifar J, Wang L, Huber I, Archontoulis SV (2021) A time-dependent parameter estimation framework for crop modeling. Sci Rep 11(1):1–15. https://doi.org/10.1038/s41598-021-90835-x
Angus JF, Nix HA, Russell JS, Kruizinga JE (1980) Water use, growth and yield of wheat in a subtropical environment. Aust J Agric Res 31:5. https://doi.org/10.1071/AR9800873
Asseng S, Bar-Tal A, Bowden JW, Keating BA, Van Herwaarden A, Palta JA, Huth NI, Probert ME (2002) Simulation of grain protein content with APSIM-Nwheat. Eur J Agron 16(1):25–42. https://doi.org/10.1016/S1161-0301(01)00116-2
Balci O (1995) Principles and techniques of simulation validation, verification, and testing. Proceedings of the 1995 winter simulation conference. doi:https://doi.org/10.1145/224401.224456
Beven K, Freer J (2001) Equifinality, data assimilation, and uncertainty estimation in mechanistic modelling of complex environmental systems using the GLUE methodology. J Hydrol 249:1–4. https://doi.org/10.1016/S0022-1694(01)00421-8
Beven K (2018) Environmental modelling: an uncertain future? CRC Press, Boca Rotan
Boote KJ, Jones JW, Pickering NB (1996) Potential uses and limitations of crop models. Agron J 88(5):704–716. https://doi.org/10.2134/agronj1996.00021962008800050005x
Bouman BAM, van Keulen H, van Laar HH, Rabbinge R (1996) The “School of de Wit” crop growth simulation models: a pedigree and historical overview. Agric Syst 52(2–3):171–198. https://doi.org/10.1016/0308-521X(96)00011-X
Brisson NC, Gary E, Justes R, Roche B, Mary D, Ripoche D, Zimmer, et al. (2003) An overview of the crop model stics’. Eur J Agron 18(3–4): 309–32. https://doi.org/10.1016/S1161-0301(02)00110-7
Brown HE, Moot DJ, Fletcher AL, Jamieson PD (2009) A framework for quantifying water extraction and water stress responses of perennial lucerne. Crop Pasture Sci 60:785–794. https://doi.org/10.1071/CP08415
Brown H, Huth N, Holzworth D (2018) Crop model improvement in APSIM: using wheat as a case study. Eur J Agron 100:141–150. https://doi.org/10.1016/j.eja.2018.02.002
Brown HE, Huth NI, Holzworth DP, Teixeira EI, Wang E, Zyskowski RF, Zheng B (2019) A generic approach to modelling, allocation and redistribution of biomass to and from plant organs. in silico Plants (1):1 https://doi.org/10.1093/insilicoplants/diy004
Brukhin V, Morozova N (2011) Plant growth and development-Basic knowledge and current views. Math Model Nat Phenom 2:1–53. https://doi.org/10.1051/mmnp/20116201
Bunce C (2009) Correlation, agreement, and Bland-Altman analysis: statistical analysis of method comparison studies. Am J Ophthalmol 148(1):4–6. https://doi.org/10.1016/j.ajo.2008.09.032
Gaber N, Foley G, Pascual P, Stiber N, Sunderland E, Cope B, Nold A, Saleem Z (2009) Guidance on the development, evaluation, and application of environmental models. Office of the Science Advisor, United States Environmental Protection Agency. EPA/100/K-09/003
Gaetani I, Pieter-Jan H, Hensen JLM (2020) A stepwise approach for assessing the appropriate occupant behaviour modelling in building performance simulation. J Build 13(3):362–377. https://doi.org/10.1080/19401493.2020.1734660
Gauch HG, Hwang JG, Fick GW (2003) Model evaluation by comparison of model-based predictions and measured values. Agron J 95:6. https://doi.org/10.2134/agronj2003.1442
Giavarina D (2015) Understanding Bland Altman Analysis. Biochem Med 25(2):141–151. https://doi.org/10.11613/BM.2015.015
He D, Wang E, Wang J, Lilley J, Lou Z, Pan X, Pan Z, Yang N (2017) Uncertainty in canola phenology modelling induced by cultivar parameterization and its impact on simulated yield. Agric for Meteorol 232:163–175. https://doi.org/10.1016/j.agrformet.2016.08.013
Hochman Z, Dang YP, Schwenke GD, Dalgliesh NP, Routly R, McDonald M, Daniells IG, Manning W, Poulton PL (2007) Simulating the effects of saline and sodic subsoils on wheat crops growing on Vertosols. Aust J Agric Res 58:8. https://doi.org/10.1071/AR06365
Holzworth DP, Huth NI, deVoil PG (2011) Simple software processes and tests improve the reliability and usefulness of a model. Environ Model Softw 26(4):510–516. https://doi.org/10.1016/j.envsoft.2010.10.014
Holzworth DP, Huth NI, deVoil PG, Zurcher EJ, Herrmann NI, McLean KC et al (2014) APSIM – evolution towards a new generation of agricultural systems simulation’. Environ Model Softw 62:327–350. https://doi.org/10.1016/j.envsoft.2014.07.009
Holzworth D, Huth NI, Fainges J, Brown H, Zurcher E, Cichota R, Verrall S, Herrmann NI, Zheng B, Snow V (2018) APSIM next generation: overcoming challenges in modernising a farming systems model. Environ Model Softw 103:43–51. https://doi.org/10.1016/j.ensoft.2018.02.002
Hornberger GM, Spear RC (1981) Approach to the preliminary analysis of environmental systems. J Environ Mgmt 12:1
Huth NI, Thorburn PJ, Radford BJ, Thornton CM (2010) Impacts of fertilisers and legumes on N2O and CO2 emissions from soils in subtropical agricultural systems: a simulation study. Agric Ecosyst Environ 136:3–4. https://doi.org/10.1016/j.agee.2009.12.016
Jones CA, Kiniry JR, Dyke PT (1986) CERES-Maize. A simulation model of maize growth and development, Texas A& M University Press, College Station
Kivi MS, Blakely B, Masters M, Bernacchi CJ, Miguez FE, Dokoohaki H (2022) Development of a data-assimilation system to forecast agricultural systems: a case study of constraining soil water and soil nitrogen dynamics in the APSIM model. Sci Tot Environ 820:3. https://doi.org/10.1016/j.scitotenv.2022.153192
Lemyre FC, Chalifoux K, Desharnais B, Mireault P (2021) Squaring things up with R2: what it is and what it can (and cannot) tell you. J Anal Toxicol 00:1–6. https://doi.org/10.1093/jat/bkab036
Liu D, Guo S, Wang Z, Liu P, Yu X, Zhao Q, Zhou H (2018) Statistics for sample splitting for the calibration and validation of hydrological models. Stoch Environ Res Risk Assess 32:3099–3116. https://doi.org/10.1007/s00477-018-1539-8
Moeller C, Pala M, Manschadi AM, Meinke H, Sauerborn J (2007) Assessing the sustainability of wheat-based cropping systems using APSIM: model parameterisation and evaluation. Aust J Agric Res 58:1. https://doi.org/10.1071/AR06186
Monteith JL (1986) How do crops manipulate water supply and demand? Philos Trans R Soc Lond Ser A 316:1537
Monod H, Naud C, Makowski D (2006) Uncertainty and sensitivity analysis for crop models. Working with dynamic crop models: evaluation, analysis, parameterization, and applications. Elsevier, Amsterdam, pp 55–100
Moriasi DN, Arnold JG, Van Liew MW, Bingner RL, Harmel RD, Veith TL (2007) Model evaluation guidelines for systematic quantification of accuracy in watershed simulations. Trans ASABE 50:3. https://doi.org/10.13031/2013.23153
Moriasi DN, Gitau MW, Pai N, Daggupati P (2015) Hydrologic and water quality models: performance measures and evaluation criteria. Trans ASABE 58:6. https://doi.org/10.13031/trans.58.10715
Morris MD (1991) Factorial sampling plans for preliminary computational experiments. Technometrics 33:161–174
Muleta MK, Nichlow JW (2005) Sensitivity and uncertainty analysis coupled with automatic calibration for a distributed watershed model. J Hydro 306:1–4. https://doi.org/10.1015/j.jhydrol.2004.09.005
Muller S, Muñoz-Carpena R, Kiker G (2011) Model Relevance. In Climate. NATO Science for Peace and Security Series C: environmental security. Springer, Dordrecht, pp. 39–65. https://doi.org/10.1007/978-94-007-1770-1_4
Nissanka SP, Karunaratne AS, Perera R, Weerakoon WMW, Thorburn PJ, Wallach D (2015) Calibration of the phenology sub-model of APSIM-Oryza: going beyond goodness of fit. Environ Model Softw 70:128–137. https://doi.org/10.1016/y.envsolft.2015.04.007
Ojeda JJ, Huth N, Holzworth D, Raymundo R, Zyskowski F, Sinton SM, Michel AJ, Brown HE (2021) Assessing errors during simulation configuration in crop models-a global case study using APSIM-Potato. Ecol Modell 458:109703. https://doi.org/10.1016/j.ecolmodel.2021.109703
Passioura JB (1996) Simulation models: science, snake oil, education, or engineering? Agron J 88(5):690–694. https://doi.org/10.2134/agronj1996.00021962008800050002x
Peake A, Hochman Z, Dalgliesh NP (2010) A rapid method for estimating the plant available water capacity of vertosols. In Australian Society of Agriculture conference, November (pp. 15-19)
Piñeiro G, Perelman S, Guerschman JP, Paruelo JM (2008) How to evaluate models: observed vs. predicted or predicted vs. observed? Ecol Modell 216:316–322. https://doi.org/10.1016/j.ecolmodel.2008.05.006
Prost L, Cerf M, Jeuffroy M (2012) Lack of consideration for end-users during the design of agronomic models. A review. Agron Sustain Dev 32, no. 2 (April 2012): 581–94. https://doi.org/10.1007/s13593-011-0059-4
Ritchie JT, Nesmith DS (1991) Temperature and crop development. Modeling plant and soil systems, Volume 31. American Society of Agronomy, Inc. Crop Science Society of America, Inc. Soil Science of American, Inc., pp. 5-29. https://doi.org/10.2134/agronmonogr31.c2
Robertson MJ, Carberry PS, Huth NI, Turpin JE, Probert ME, Poulton PL, Bell M, Wright GC, Brinsmead YSJ, RB, (2002a) Simulation of growth and development of diverse legume species in APSIM. Aust J Agric Res 53:4. https://doi.org/10.1071/ar01106
Robertson MJ, Asseng S, Kirkegaard JA, Wratten N, Holland JF, Potter TD, Burton W, Walton GH, Moot DJ, Wratten N, Farre I, Asseng S (2002b) Environmental and genotypic control of time to flowering in canola and Indian mustard. Aust J Agric Res 53:7. https://doi.org/10.1071/ar01182
Robertson MJ, Lilley JM (2016) Simulation of growth, development and yield of canola (Brassica napus) in APSIM. Crop Pasture Sci 67:332–344. https://doi.org/10.1071/CP15267
Saltelli A (2002) Sensitivity analysis for importance assessment. SRA 22:3. https://doi.org/10.1111/0272-4332.00040
Saltelli A (2019) A short comment on statistical versus mathematical modelling. Nat Commun 10:3870. https://doi.org/10.1038/s41467-019011865-8
Seidel SJ, Palosuo T, Thorburn P, Wallach D (2018) Towards improved calibration of crop models–where are we now and where should we go? Eur J Agron 94:25–35. https://doi.org/10.1016/j.eja.2018.01006
Snowling, SD, Kramer JR (2001) Evaluating modelling uncertainty for model selection. Ecol Modell 138(1–3) (March 2001): 17–30. https://doi.org/10.1016/S0304-3800(00)00390-2
Sobol IM (2001) Global sensitivity indices for nonlinear mathematical models and their Monte Carlo estimates. Math Comput Simul 55:271–280. https://doi.org/10.1016/S0378-4754(00)00270-6
Stirling A (1999) On science and precaution in the management of technological risk: an ESTO project report. European Commission-JRC Institute Prospective Technological Studies, Seville
van Ittersum MK, Leffelaar PA, van Keulen H, Kropff MJ, Bastiaans L, Goudriaan J (2003) On approaches and applications of the Wageningen crop models. Eur J Agron 18:201–234. https://doi.org/10.1016/S1161-0301(02)00106-5
Wallach D (2006) Evaluating crop models. Working with dynamic crop models. Elsevier, Amsterdam, pp 11–53
Wang E, Robertson MJ, Hammer GL, Carberry PS, Holzworth D, Meinke H, Chapman SC, Hargreaves JNG, Huth NI, McLean G (2002) Development of a generic crop model template in the cropping system model APSIM. Eur J Agron 18:121–140. https://doi.org/10.1016/S1161-0301(02)00100-4
Whish JPM, Thompson JP, Clewett TG, Lawrence JL, Wood J (2014) Pratylenchus thornei populations reduce water uptake in intolerant wheat cultivars. Field Crop Res 161:1–10. https://doi.org/10.1016/j.fcr.2014.02.002
Yang JM, Yang JY, Liu S, Hoogenboom G (2014) An evaluation of the statistical methods for testing the performance of crop models with observed data. Agric Syst 127:81–89. https://doi.org/10.1016/j.agsy.2014.01.008
Yin X, Kropff MJ, McLaren G, Visperas RM (1995) A nonlinear model for crop development as a function of temperature. Agric for Meteorol 77:1–2. https://doi.org/10.1016/0168-1923(95)02236-Q
Zhang L, Dawes WR, Walker GR (2001) Response of mean annual evapotranspiration to vegetation changes at catchment scale. Water Resour 37:3. https://doi.org/10.1029/2000WR900325
Zhao G, Bryan BA, Song X (2014) Sensitivity and uncertainty analysis of the APSIM-wheat model: interactions between cultivar, environmental, and management parameters. Ecol Model 279:1–11. https://doi.org/10.1016/j.ecolmodel.2014.02.003
Acknowledgements
We thank the APSIM Initiative for making the APSIM software and code publicly available. We also thank Drs. Kirsten Verburg, Julianne Lilley, and Lucy Watt for conducting an internal review of the manuscript.
Funding
This work was supported by Grains Research & Development Corporation (GRDC) grant no. ULA1906-002RTX.
Author information
Authors and Affiliations
Contributions
All authors contributed to the conceptualization, design, and editing of this manuscript. H.P., H.B., D.H., J.W., and N.L. provided input in terms of approaches to model construction and testing. L.B. provided input in terms of model usage and funding acquisition. All authors provided insight about dataset acquisition and deductive approaches. H.P. wrote the original manuscript. H.B., D.H., J.W., L.B., and N.L. reviewed and commented on drafts of the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Pasley, H., Brown, H., Holzworth, D. et al. How to build a crop model. A review. Agron. Sustain. Dev. 43, 2 (2023). https://doi.org/10.1007/s13593-022-00854-9
Accepted:
Published:
DOI: https://doi.org/10.1007/s13593-022-00854-9