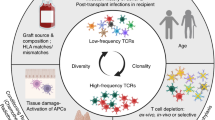
Abstract
Acute graft versus host disease (aGVHD) is a rare, but severe complication of liver transplantation (LT). It is caused by the activation of donor immune cells in the graft against the host shortly after transplantation, but the contributing pathogenic factors remain unclear. Here we show that human T cell lymphotropic virus type I (HTLV-1) infection of donor T cells is highly associated with aGVHD following LT. The presence of HTLV-1 in peripheral blood and tissue samples from a discovery cohort of 7 aGVHD patients and 17 control patients is assessed with hybridization probes (TargetSeq), mass cytometry (CyTOF), and multiplex immunohistology (IMC). All 7 of our aGVHD patients display detectable HTLV-1 Tax signals by IMC. We identify donor-derived cells based on a Y chromosome-specific genetic marker, EIF1AY. Thus, we confirm the presence of CD4+Tax+EIF1AY+ T cells and Tax+CD68+EIF1AY+ antigen-presenting cells, indicating HTLV-1 infection of donor immune cells. In an independent cohort of 400 patients, we verify that HTLV-1 prevalence correlates with aGVHD incidence, while none of the control viruses shows significant associations. Our findings thus provide new insights into the aetio-pathology of liver-transplantation-associated aGVHD and raise the possibility of preventing aGVHD prior to transplantation.
Similar content being viewed by others
Introduction
Graft-versus-host disease (GVHD) is defined as immunological damage to host tissues and organs due to the activation of donor immune cells in graft by the host’s major histocompatibility complex (MHC)1,2. It is a type of rejection that is stronger than typical rejections, destroying the skin and mucosal barriers of the host and leading to severe infections and ultimately lethal septic shock. The disease progression can be less than two weeks from onset to mortality3. Generally, GVHD can be categorized into two types, namely acute (aGVHD) and chronic (cGVHD), depending on whether the disease manifests before or after the 100th day since transplantation4. In contrast to the high incidence (20–80%) of aGVHD in patients receiving hematopoietic stem cell transplantation (HSCT)5,6, the incidence is much lower for solid organ transplantations, such as liver transplantation (LT) (0.1–2%)7. However, the mortality of LT patients with aGVHD is as high as 80–100%8, whereas the overall lethality of transplant recipients is 9.4% within one year9. The low incidence of GVHD with high mortality is presented in all kinds of solid organ transplantations. However, there is still a lack of theory to explain this issue10.
More than 20 years ago, donor-derived T cells were identified as the primary cause of GVHD11. The introduction of massive donor-derived T cells contributes to the high incidence of GVHD in HSCT patients5,6, whereas LT patients receive a tiny proportion of donor immune cells. When a liver graft is implanted, 109−1010 donor-derived immune cells are transferred into the recipient’s body, including T cells, B cells, natural killer (NK) cells, and monocytes12. In some cases, these cells can be detected in the recipient’s peripheral blood and bone marrow between 3 and 100 days after surgery which matches the onset time of aGVHD13. Any transplant that transfers viable allogeneic lymphocytes into the recipient carries the potential risk of GVHD, such as transfusion-associated GVHD14,15.
The critical process of the development of aGVHD is the activation of T-cell receptors (TCR) on donor-derived T cells by the antigen-presenting cells (APC) with corresponding host MHC or minor histocompatibility antigen (miH) peptides16. Donor CD8+ T cells alone can induce GVHD by identifying disparities of MHC class I molecules, while CD4+ T cells induce GVHD by recognizing MHC class II molecules10,17,18. T cells are the main effectors causing cell death in target tissues, mediated by various cytotoxic effects10. As a pathogenic and T-cell-tropic human retrovirus, human T-cell lymphotropic virus type I (HTLV-1) can infect and immortalize CD4+ T cells19,20. In different animal models of HTLV-1 infection, established in macaques, rabbits, and humanized mice, the virus can cause lethal CD4+ T-cell expansion21,22,23. Of note, the bone marrow-liver-thymus (BLT) mouse model was prone to aGVHD, related to elevated levels of HTLV-1 RNA in blood21. HLA matching is performed before HSCT to avoid massive donor-derived T cells attacking the hosts, while HLA matching is not mandatory in liver transplantations. Generally, the proportion of donor lymphocytes in LT is too small to induce aGVHD even though HLA is mismatching between the donors and the recipients. Additionally, HLA miss-matching did not correlate with LT outcome or rejection rate24,25. In HSCT, tissue damage and GVHD seem to be the reciprocal causation26,27,28. However, in LTs, without tissue damage caused by conventional chemotherapy, the risk factors that trigger and induce the expansion of donor-derived T cells for aGVHD development remain unknown, which is our focus.
In this study, we hypothesize that viral infection plays a role in expanding donor-derived T cells in LT, which might have been masked by the severe fungi and bacteria infections in the course of aGVHD. From 2015 to 2020, seven aGVHD patients that were DCD (donor after circulatory death) recipients had been identified among 3763 liver transplant recipients in our center. Therefore, the incidence of aGVHD is 0.18% (7/3,763). To reveal the cryptogenic infections associated with developing aGVHD, we design a cocktail of probes targeting eight non-hepatotropic viruses. As a result, HTLV-1 infections are explicitly found in all the seven aGVHD patients. Furthermore, the multiplex immunohistology (IMC) results confirm that donor-derived CD4+ T cells are increased by HTLV-1 infection. Additionally, joint analysis of mass cytometry (CyTOF) and IMC results suggest the aGVHD-specific Tax+CD68+ APCs take up HTLV-1 and transmit the virus and immune signals. Thus, our study indicates that HTLV-1 is involved in the entire process of aGVHD.
Results
HTLV-1 only found in aGVHD patients after liver transplantation
To reveal the cryptogenic infections associated with the development of aGVHD, TargetSeq was adopted to detect viral infection in samples from two patients (ID139 and ID141) in the aGVHD group and 17 recipients in the control group (Fig. 1A). Detailed patient information and grouping principles were described in the Method and materials (Table S1). In the aGVHD group, the peripheral blood, skins, or liver biopsy tissues from only two patients were acquired due to limits of sample collection (Fig. 2A). Likewise, peripheral blood and disease-related lesion tissue specimens, including liver tissues, infectious site tissues, and lung metastases of liver tumors, were collected from the control group (Fig. 2A and Table S1). TargetSeq allowed us to determine the existence of viruses (HHV-6A, HTLV-1, HTLV-2, HTLV-4, EBV, adenovirus, CMV, and parvovirus-1), especially whether viruses were in the latent infection phase29. Latent infection, where viral DNA had been integrated into the host genome, is a way for viruses to achieve persistent and lifelong infection in humans and participate in the pathogenesis of various complications30. In both the aGVHD and control groups, HTLV-2 and HTLV-4 were not detected and therefore excluded from subsequent analysis (Fig. 2A). HHV-6A and HTLV-1 were detected in all the aGVHD samples tested, while EBV and adenovirus were detected in some of the aGVHD samples (Fig. 2A). Viral host gene integrations of HTLV-1, HHV-6A, and adenovirus were found in both the liver biopsy tissues and the skin tissues from patient 139, and EBV integrations in liver biopsy tissues only. Meanwhile, viral host gene integrations of HTLV-1, HHV-6A, and EBV were detected in only the liver biopsy tissues of patient 141 (Fig. 2A, B). The presence of viruses was compared between groups by Fisher’s exact test and corrected with Benjamini-Hochberg adjustment (Fig. 2C). Compared with the control group, the percentage of HTLV-1 positive patients was significantly higher in the aGVHD group (adjusted p value = 0.0406) (Fig. 2C). Parvovirus-1 and CMV were not detected in any of the aGVHD samples. In addition, HTLV-1 was not detected in the peripheral blood or tissue samples from the control patients, which contrasts with the aGVHD group where this virus was detected in all the samples (Fig. 2A, C, D). Although HHV-6A was detected in all samples from the aGVHD group, HHV-6A was also present in the control group. There was no significant difference in the HHV-6A infection rates between the aGVHD and control groups (Fig. 2A, C). Therefore, these results preliminarily indicated the association between HTLV-1 infection and the occurrence of aGVHD. In addition, some of the viruses were not detected in the peripheral blood of the patients with tissue infections, which suggests it may be more reliable to perform virus screening on both tissue and peripheral blood (Fig. 2D).
A Multiple viral DNA and RNA sequences were detected in 37 specimens from the patients in the aGVHD group (N = 2) and the control group (N = 17). Tested specimens included skins with rash, peripheral blood, liver grafts, intestines, ventricular walls, and lungs. B CyTOF was performed on a total of 16 samples of peripheral blood mononuclear cells (PBMCs) from the patients in the aGVHD group (N = 3) and the control group (N = 10). C 17 formalin-fixed and paraffin-embedded (FFPE) samples from the patients in the aGVHD group (N = 7), including seven skin tissues, five native liver tissues, three graft liver tissues, and two bone marrow tissues, were assessed with IMC.
A The heatmap depicts the detection of virus or virus/host integration in each sample from two patients in the aGVHD group and 17 patients in the control group. Negative, positive, and host integration results are marked as blue, yellow, and red, respectively. B Patient numbers of virus infection and host integration were counted. C The percentages of virus infection rates in each group were calculated and compared between the aGVHD group (N = 2) and control group (N = 17). All p values were calculated using Fisher’s exact test and corrected with Benjamini-Hochberg adjustment. *p = 0.0406. D The percentage of patients with virus detected in either peripheral blood or tissue samples. E The heatmap depicts basic clinical information of the 400 patients in the validation cohort, which includes age, body mass index (BMI), sample collection time, pre-operative diagnosis, and concomitant diseases. The two HTLV-1 infection patients are marked by red arrows with annotations of the viral loads. (Benign ESLD: benign end-stage liver disease, Malignant LD: malignant liver disease, Complications after LT: complications after liver transplantation, NC: no concomitant disease.).
To further investigate the prevalence of HTLV-1 in our center and verify our hypothesis of HTLV-1 infection and aGVHD process, an independent cohort of 400 patients, including liver surgery patients, recipients, and donors of liver transplants (Table S2), were recruited from January 2021 to June 2021 for RT-PCR detection of HTLV-1 infection. A total of two patients were found HTLV-1 positive, including one donor of transplantation and one patient after partial hepatectomy (Fig. 2E). Notably, the recipient of the HTLV-1 positive donor was not infected due to the viral load of the donor being lower than the lowest load that can cause viral infection (9 × 104)31. The other HTLV-1-positive patient with co-infection with hepatitis C died on the second day after surgery for acute liver failure. None of the 400 patients in this cohort was diagnosed with aGVHD, and there was no HTLV-1 positive recipient. Therefore, the prevalence of HTLV-1 in our center in patients without aGVHD was 0.5%, with a 95% confidence interval of [0.01%, 1.93%], which matched the reported prevalence of 0.1% to 1%32. Given that, the possibility that the co-occurrences of the two rare diseases were a coincidence was less than 10−4 using a binomial probability calculation, suggesting that HTLV-1 infection should be closely related to aGHVD development after LT.
aGVHD-specific circulating immune cells (Tax+CD68+) uncovered in peripheral blood with features of APCs
Since the relationship between HTLV-1 and aGVHD is statistically supported, a series of explorative experiments were conducted to uncover the underlying biological mechanisms further. Mass cytometry (CyTOF) was adopted for systematic and deep phenotyping of the PBMCs from the aGVHD (N = 3) and control (N = 10) patients (Fig. 1B). Samples were collected from aGVHD patients at two different time points, and once from control patients. A CyTOF panel, including 31 metal isotope-tagged antibodies (Table S4), was designed to acquire a global overview of the leukocytes in peripheral blood. The panel contained 30 lineage markers to distinguish the significant leukocyte subsets and a marker to trace HTLV-1 infection at the cellular level. Mainly, a metal-labeled antibody against Tax protein, an HTLV-1 specific RNA transcription enhancer33,34, was included in the panel. To characterize the phenotypes of immune cell subsets, t-distributed stochastic neighbor embedding analysis was used to generate a two-dimensional map in Cytofkit35. For each sample, at least 5 × 103 leukocytes (CD45+ cells) were selected for subsequent data analysis (Fig. S1). Results showed that the Tax protein signal was positive in all six aGVHD samples while negative in all 10 samples from the control group. The positive/negative ratio of the signal intensities was up to 50 (Fig. 3A).
A The two histograms visualize the intensities of HTLV-1-specific Tax signal in CD45+ populations (Fig. S1) from the two groups of PBMC samples. In aGVHD group, PBMCs from three aGVHD patients were collected at two-time points, T1 and T2, between onset and death. Ten patients in the control group received liver transplants and their PBMCs were collected at one postoperative time point when the complication occurred. PBMCs from aGVHD patients show high levels of HTLV-1 Tax expression. B t-SNE dimensional reduction was performed on CD45+ cells to explore the heterogeneity across patients. The color scale indicates the arcsinh-transformed signal intensity of Tax. The clusters with positive Tax expressions are marked through manual gating (red circles), primarily presented in the aGVHD group. C PhenoGraph and t-SNE depict the clustering result with 30 clusters identified automatically. The aGVHD group displays distinct cluster distributions from the control group. The Tax+ subpopulations are marked with hollow red circles (Table S6). D A heatmap depicts the median expression of cell markers in the seven Tax+ subpopulations. The HLA-DR+ subpopulations are marked by a hollow red rectangle. The color bars indicate arcsinh-transformed signal intensities of the proteins. E Percentages of the seven Tax+ subpopulations in CD45+ cells were compared between the aGVHD and control groups. Tax+ cells are presented mainly in the aGVHD group. (Intermediate monocyte: iMo; non-classical monocyte: ncMo; Conventional dendritic cell: cDC; CD8+ T cell.).
In Fig. 3B, every dot represents a single cell, and color denotes the expression level of Tax protein. The spatial distributions of the immune cells were different between the aGVHD and control groups, suggesting different immune responses in these patients (Fig. 3B). We then clustered immune cells into 30 clusters based on the surface marker expressions with PhenoGraph35 (Fig. 3C). The major different clusters between the aGVHD and control groups were Tax+CD68+ clusters (Fig. 3B and S2). In accordance with the result of TargetSeq that HTLV-1 infection should be closely related to aGHVD development after LT, Tax+ cells were only detected in cells from the aGVHD group, which composed the clusters 22–26# and 29-30# (Fig. 3B, C). The cell phenotypes of the Tax+ clusters are listed in Table S6. Cluster 23#, 25#, 29# and 30# were classified as CD3−CD19−CD11c+ CD123− cells. Among them, Cluster 29# were HLA-DR+ cells, which can be defined as conventional dendritic cells (cDC cells). Dendritic cells (DC), constituting the mononuclear phagocyte system (MPS), are considered to be the most potent APCs36. Because CD68 is a selective marker for human monocytes and macrophages, Clusters 22#, 23#, and 30# (CD3−CD19−CD14−CD16+HLA-DR−CD68+) were considered non-classical monocytes37. Cluster 25# was intermediate monocytes (CD3−CD19−CD14+CD11c+CD16+CD68+). Cluster 24# (CD3−CD19−CD14−CD16−CD11c−CD123−HLA-DR−CD68+) expressed activation markers such as CD3838 and CD6939, indicating these cells were highly activated. Meanwhile, cluster 26# (CD19−CD14−CD3+CD8+Tax+) was characterized as Tax+CD8+T cells (Fig. 3C, D, and S2). PhenoGraph clustered 10 to 100 times more cells from the aGVHD group than the control group into the seven Tax+ clusters (Fig. 3E). The percentages of the clusters 22#, 26#, and 29–30# exceeded 5%, occupying high proportions in leukocytes in the aGVHD samples. DCs, monocytes, and macrophages have long been identified as antigen-presenting cells in circulation and in tissues40,41,42. APC, reacted with antigen, presented MHC II groove-bound peptides to CD4+ T cells that share the same allele, provoking immune responses43. Our CyTOF results revealed aGVHD-specific immune cell subsets, especially Tax+CD68+ APCs.
“Triple-positive T cells” confirmed to be donor-derived T cells in skin sections
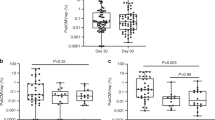
Following the analysis of Tax+ immune cell subsets in peripheral blood, IMC was further applied to study the spatial distribution of the Tax+ immune cells in the skin lesions, which is one of target organs in aGVHD (Fig. 1C). An IMC panel of 19 metal isotope-tagged antibodies was designed to acquire a global overview of the immune cells in the skin lesions and reveal the interactions between HTLV-1 and immune cells (Table S5). The panel contained two structure protein markers, 13 immune cell markers, an HTLV-1 specific marker (Tax protein), and a Y chromosome marker to track the source of cells. IMC was performed on skin lesions from all seven patients in the aGVHD group. Among them, 4 female patients (ID139, ID141, ID143, and ID144) received livers from male donors. Thus, eukaryotic translation initiation factor 1 A Y-linked (EIF1AY), expressed by the corresponding gene located on Y chromosome, was targeted to track donor-derived cells in samples from female recipients44. The signals of EIF1AY were shown on samples from female samples (Fig. 4A–D) but not on male samples (Fig. 4E–H). In addition, blood serum from 2 aGVHD patients (ID144 and ID145) and 2 control patients were also subjected to ELISA test, and the HTLV-1 antibody titers were 30 to 100-fold higher in the aGVHD samples than in the control samples (Fig. S3).
Representative IMC images of two independently stained skin tissues from 7 aGVHD patients show the overlap of Tax (red), DNA (blue) and different immune response markers. Displayed channels for each column were: A Tax (red)/DNA (blue); B CD4 (green)/Tax (red)/EIF1AY (white)/DNA (blue); C C4d (green)/Tax (red)/EIF1AY (white)/DNA (blue); D CD68 (green)/Tax (red)/EIF1AY (white)/DNA (blue). E Tax (red)/DNA (blue); F CD4 (green)/Tax (red)/DNA (blue); G C4d (green)/Tax (red)/DNA (blue); H CD68 (green)/Tax (red)/DNA (blue). Red arrows pinpoint the multi-signals overlap position and red boxes show the magnified view of CD4+T cells with Tax signal. Scale bar = 100 μm for landscape view and scale bar = 20 μm for magnified view.
The interactions between various immune cells and HTLV-1 in skin tissue sections are shown in Fig. 4. The region of interest (ROI) was defined as the lymphocyte-rich area in the epidermal–dermal junction selected by optical microscopy. In the specimen sections from every patient in the aGVHD group, Tax protein signals were detected in the nucleus, cytoplasm, and the extracellular space (secreted protein), confirming the presence of the HTLV-1 viral protein in targeted organs, including skin and liver. In addition, Fig. 4B shows the overlay of CD4, EIF1AY, and Tax protein signals. The triple overlap signals can be found in all the skin sections from female recipients (Fig. 4B–D). The Tax+CD4+ cells also expressed CD3, so Tax+CD4+ cells were defined as CD4+ T cells (Fig. S4, S5). The overlay of CD4, EIF1AY, and Tax protein signals revealed that the effector T cells of aGVHD was “triple-positive T cells”, which were donor-derived and HTLV-1-infected CD4+ T cells. Fig. 4C, G demonstrate abundant C4d expression in immune cells that infiltrated skin lesions and show co-expression of Tax and C4d protein. Activated by the antigen-antibody reaction, C4d is an important marker in antibody-mediated humoral rejection after organ transplantation and also been reported to present after the occurrence of GVHD45,46,47. It might be a sign of interaction between viral infection and immune rejection during aGVHD progression48. Additionally, the overlapped signals of CD68, Tax, and EIF1AY in Fig. 4D, H implied the presence of donor-derived APCs, which is consistent with published literatures49,50,51. Furthermore, our results indicate that Tax expression was not detected in CD8+ T cells or B cells in the skin (Fig. S6). Single signal images of the 12 lineage markers were presented in Fig. S4, S5, S7, and S8. Thus, by labeling EIF1AY and Tax, IMC results reproduced the interaction between HTLV-1 and other immune cells, especially donor-derived cells, during the pathogenesis of aGVHD.
To fully display the HTLV-1 detection result assessed with multiple methods in the discovery cohort, the positive rate and specific detection information of HTLV-1 are listed in Table 1 and Table S1. By tracking the clinical records, we integrated the PCR detection results of HHV-6A, EBV, adenovirus, CMV, and parvovirus-1 in all the 24 patients in the discovery cohort and performed statistical analysis between the two groups (Table 1). By Fisher’s exact test and corrected with Benjamini–Hochberg adjustment, significant differences in positive rate of HTLV-1 detection were found between the control and aGVHD groups (adjusted p value <0.0001). The four methods cross-validated that all and only the seven aGVHD patients were infected with HTLV-1.
CD68+ mononuclear phagocyte subsets defined in skin sections of aGVHD patients
After identification of Tax+ CD68+ cells in peripheral blood and skin sections, the characteristics of APCs in skin lesions of all aGVHD patients were investigated by IMC to further reveal the role of HTLV-1 infection in aGVHD. The primary APCs in the skin are tissue mononuclear phagocyte (MNP) subsets, which are two prominent families, DCs and tissue-resident macrophages40. DCs are further divided into conventional DC (cDC) and plasmacytoid DC (pDC). cDCs, pDCs, monocyte-derived macrophages (MDM), and tissue-resident macrophages respectively express CD11c+HLA-DR+CD68+, CD123+HLA-DR+CD68+, CD14+CD11c−CD68+, and CD14-CD11c−CD68+ cells52,53. The phenotypes and distributions of the four MNPs found by IMC in the skin lesions were presented by three representative patients (ID139, ID140, and ID145, Fig. 5A–C). Single color images of HLA-DR, CD14, CD123, CD68, and CD11c from the seven GVHD patients were shown with nuclear signals in Fig. S7 and S8. The signals of DNA, HLA-DR, CD14, CD123, CD68, CD11c, and the merged panels of these signals are shown in Fig. 5A–C. The summarized MNP phenotypes of all seven aGVHD patients are shown in Fig. 5D. Macrophages existed in skin sections of all the seven aGVHD patients, and MDMs were found in four patients (ID139, ID141, ID 142, ID144), while cDCs and pDCs only presented in skin sections of two patients (ID140 and ID143). In addition, some of the MNPs expressed the Tax protein (Fig. 4D, H), suggesting that they take up HTLV-1 and transmit the virus and the signal outward. Thus, MNP cells found in aGVHD patients were diverse, mainly DCs and macrophages, and some of them were Tax+CD68+ cells, consistent with the aGVHD-specific cell subsets in peripheral blood. The IMC results of skin sections reproduced the antigen-presenting responses and presented the APCs which were previously detected in peripheral blood and possibly migrated into the tissues.
Representative IMC images of two independently stained skin tissues from 3 aGVHD patients (ID139, 140, and 145) show the seven single signals of A DNA (blue), HLA-DR (red), CD11c (purple), CD14 (yellow), B CD123 (light blue), CD68 (green). C overlay signal of seven markers. The same markers are shown in images in each column. Red, yellow, green, and blue arrows, respectively, pinpoint the monocyte-derived macrophages (MDM), macrophages, plasmacytoid dendritic cells (pDC), and conventional dendritic cells (cDC). Red, yellow, green, and blue boxes respectively show the magnified multi-signal overlap position of MDMs, macrophages, pDC, and cDC. Scale bar = 100 μm for landscape view and scale bar = 20 μm for magnified view. D Each column is mononuclear phagocyte (MNP) identification in skin sections from each patient. Each row is the comprehensive phenotyping of one MNP, and the symbol+/− indicates the MNP is existence/absent in skin sections of each patient. Displayed channels for each row are: macrophage (CD14−CD11c−CD68+); MDM (CD14+CD11c−CD68+); cDC (CD11c+HLA-DR+CD68+); pDC (CD123+HLA-DR+CD68+).
HTLV-1 specific signals in liver sections indicated HTLV-1 was either donor-derived or pre-existed in the recipients
To trace the source of HTLV-1, we further performed IMC to detect Tax protein expression in the liver graft and native liver of recipients. In the liver biopsies (liver graft) from female patients who received the organ from male donors (ID139), the Tax protein was detected in CD68+ EIF1AY+ cells, which confirmed donor-derived HTLV-1 infection from the liver graft (Fig. 6A, 4D and S9). In the native liver (ID139, ID144, and ID145), the Tax protein was detected on CD68+ cells in the native liver section, indicating the presence of pre-existing HTLV-1 infection in these patients (Fig. 6A). For both patients ID140 and ID141, the Tax protein signal was positive on the liver grafts (Fig. 6A) but negative on the native livers (Fig. 6B), suggesting HTLV-1 infection after surgery, probably donor-derived. HTLV-1 infection was not identified in the bone marrow sections or other liver tissues (Fig.6B). These results preliminarily revealed the viral infection pathways, either donor-derived or recipient pre-existed.
A Representative Tax+ images of two independently stained liver graft and native liver show two overlap patterns. Each row of figures display the same tissue type. Displayed channels for liver graft samples from left to right are: CD68 (green)/Tax (red)/EIF1AY (white)/DNA (blue), CD68 (green)/Tax (red)/DNA (blue), CD68 (green)/Tax (red)/EIF1AY (white)/DNA (blue). Displayed channels for native liver samples are CD68 (green)/Tax (red)/DNA (blue). Red arrows pinpoint the multi-signal overlap position. B Representative Tax− IMC images of two independently stained bone marrow and native liver sections show an overlap of Tax (red) and DNA (blue). Scale bar = 50 or 100 μm, shown in the images, respectively.
Discussion
In this study, we detected HTLV-1 cDNA and HTLV-1 specific Tax proteins from peripheral blood samples, fresh tissue sections, and formalin-fixed and paraffin-embedded (FFPE) tissue sections of all aGVHD-diagnosed patients by pre-synthesized TargetSeq, CyTOF, and IMC. The association between HTLV-1 infection and the occurrence of aGVHD has been determined by statistical analysis. Furthermore, CyTOF uncovered aGVHD-specific circulating Tax+CD68+ immune cells in peripheral blood, including monocytes and DCs. Integrated analysis of CyTOF and IMC results reproduced the antigen-presenting responses during aGVHD after LT. Some MNPs, mainly DCs, MDMs, and macrophages, in the skin co-expressed Tax and CD68 proteins. It explains the migration of APCs from peripheral blood to skin lesions, during which they took up HTLV-1 and transmitted the viruses and immune signals. Most importantly, we discovered the pathogenic host-targeting T cells by IMC, which were “triple-positive T cells”. Our results confirmed the existence of donor-derived CD4+ T lymphocytes in targeted recipient organs, which were possibly expanded by HTLV-1 infection, as a potential initiator of aGVHD after LT. These findings associate the incidence of aGVHD with a viral infection, providing new perspectives for its prevention and treatment.
HTLV-1 is a highly pathogenic virus similar to HIV, but due to its geographic distribution and long latency period, HTLV-1 has not been adequately heeded20. In contrast to HIV causing T-cell death, HTLV-1 can efficiently infect T cells, sustain their growth, and immortalize them through Tax protein-inducing kappa B-specific proteins19,20. Expression of the HTLV-1 Tax protein induces enhancement of T-cell promoting factors, IL-2, IL-4, IL-6, T-cell telomerase, and Tax/rex mRNA, and reduction of T-cell apoptotic factors, such as Bim (a pro-apoptotic protein), leading to excessive proliferation of T cells (generally CD4+ T-cell) and unlimited expression of the IL-2R alpha and IL-2 genes19,20. Unlimited proliferation of T cells, caused by HTLV-1 infection, acts on the human immune system and becomes an enhancer of rejection54,55. Therefore, HTLV-1 virus infection may expand T cells, possibly for both hosts and donors, resulting in a potential tendency for host-to-graft and graft-to-host rejections.
The confirmation and prevention of HTLV-1 infection depend on the perioperative preparation of transplantation. Viral infection can occur when the number of cells containing the provirus is more than 9 × 104 31. Due to the low infection rate and geographical limitations of HTLV-1, many transplantation centers do not routinely screen for HTLV-1, resulting in overlooked viral infection. In our study, expansion of donor T cells occurred regardless of the HTLV-1 infection source (donor-derived, pre-existing, or transfusion). The lethal CD4+ T cells expansion is supposed to underlay aGVHD onset and progression. Therefore, our data suggest that donors, recipients, and blood supply sources should undergo strict testing for HTLV-1 infection before surgery to prevent the disordered proliferation of T cells in organ recipients. In this study, the specificity of HTLV-1 virus infection in aGVHD patients after LT was preliminarily revealed, so a more extensive multicenter study to examine the relationship between HTLV-1 and aGVHD will be beneficial. Simplified strategies for diagnosis of HTLV-1 infection and antiretroviral therapy will play positive roles in the prevention and treatment of aGVHD after LT. At the same time, it explains one of the various viral hazards in humans and calls for more research on virus-related lymphocytic diseases.
Methods
Ethics, grouping principles, and human sample harvest
This study was approved by the ethical review board of Renji Hospital, Shanghai Jiao Tong University School of Medicine (clinical trial registration number: KY2019074). All patient samples were obtained with informed consent under the supervision of IRB.
The DCD recipients were divided into two groups, the aGVHD and control groups. The aGVHD group consisted of seven patients diagnosed with aGVHD in different years between 2015 and 2020. The control group consisted of 17 recipients, including post-transplant rejection patients (N = 4), post-transplant infection patients (N = 3), post-transplant regular recovery recipients (N = 4), and pre-operative patients (N = 6). For patients with aGVHD, post-transplant rejection, or post-transplant infection, samples were collected during the disease progression. Specimens were collected before surgery from pre-operative control patients. The patient IDs, clinical information, and groupings for the patients evaluated in this study are listed in Table S1. The independent cohort of 400 patients, including liver surgery patients, recipients, and donors of liver transplants, was recruited from January 2021 to June 2021 (Table S2).
For TargetSeq assay, fresh samples, including skin affected by a rash, peripheral blood, and biopsy tissues of livers, intestines, ventricular walls, and lungs, were harvested from 2 patients in the aGVHD group (ID139 and ID141) and 17 recipients in the control group (Table S1).
For CyTOF and subsequent analysis, PBMC samples were collected from 3 aGVHD patients (ID139, ID 140, and ID 141) and 10 recipients in the control group, including post-transplant rejection patients (N = 4), post-transplant infection patients (N = 3), and post-transplant regular recovery recipients (N = 3). The PBMCs of the aGVHD group were collected at two different time points between onset and death. The PBMCs in the control group were collected only once.
The specimens for IMC were FFPE sections of native livers, skin lesions, liver biopsies (graft tissues), and bone marrows obtained from the 7 aGVHD patients (Table S1). The FFPE sections of native livers were collected during transplant surgeries, and the other specimens were collected during the progression of aGVHD.
Detection of non-hepatotropic viruses by TargetSeq
Fresh samples (N = 37) were harvested from the aGVHD and control group patients, including skin lesions, peripheral blood, and biopsy tissues of livers, intestines, ventricular walls, and lungs (Table S1). The RNA was extracted by Pre-NAT Full-Automatic System (PerkinElmer, Massachusetts, USA). Capture probes targeting non-hepatotropic viruses (TargetSeq, iGeneTech, Beijing, China) were RNA probes based on liquid-phase chips (Fig. 1A). The targeted viruses included human herpesvirus 6 A (HHV-6A), Epstein-Barr virus (EBV), adenovirus, parvovirus-1, cytomegalovirus (CMV), HTLV-1, HTLV-2, and HTLV-4. Probe panels were designed for target region sequencing based on each virus’ whole-genome sequence acquired from the NCBI GenBank database (Table S3). For RNA viruses, the probes were designed using cDNA synthesized by reverse transcription as templates. TargetSeq was capable of capturing the full-length sequence of the target viruses and the corresponding genome integration regions.
Detection of HTLV-1 by RT-PCR
The peripheral blood was collected and the RNA was extracted by Pre-NAT Full-Automatic System (PerkinElmer, Massachusetts, USA). Real-time RT-PCR assays for HTLV-1 RNA detection were performed using Human T-cell Lymphoma Virus 1 (HTLV-1) Probe qRT-PCR Kit (YaJi Biological, Shanghai, China) in an ABI 7500 real-time PCR system (Thermo Fisher Scientific, California, USA) following the kit instruction. Each reaction mixture contained 10 μL buffer, 2 μL enzyme mixture, 2 μL mixture of forward and reverse primer, 1uL probe solution, and 5 μl specimen. The thermal cycling parameters were 30 min at 50 °C for reverse transcription, 10 min at 94 °C for predegeneration, and 40 cycles containing 15 s at 94 °C and 1 min at 60 °C.
Antibody preparation
The antibodies used in this study and the corresponding manufacturers and concentrations are listed in Table S4 and Table S5. Metal-conjugated antibodies were purchased or prepared using a Maxpar ×10 antibody labeling kit (Fluidigm Sciences, San Francisco, USA). After conjugation, antibodies were stored in Candor PBS Antibody Stabilization Solution (Candor Bioscience, Wangen, Germany) at 4 °C.
Preparation and staining of PBMCs for mass cytometry detection
PBMCs were separated by density gradient centrifugation with Ficoll-Paque PLUS (Amersham Biosciences, Piscataway, USA) (Fig. 1B). After lymphocyte extraction, cisplatin (5 μM) was used to treat cells in suspension, and the cells were then fixed in paraformaldehyde (PFA; final concentration 1.6%). Cells were stained with a cocktail of 30 metal isotope-conjugated antibodies against surface proteins (Table S4). After surface-protein staining, the cells were treated with the Transcription Factor Permeabilization kit (eBioscience, Santiago, USA) and then stained with anti-Tax antibody at room temperature for 30 min (Table S4). Nuclear were stained with 1 mL of 1:4000 diluted 191Ir/193Ir DNA intercalator (Fluidigm Sciences, San Francisco, USA) with Maxpar Perm-S Buffer (Fluidigm Sciences, San Francisco, USA) overnight at 4 °C.
Preparation of aGVHD tissue sections for IMC detection
Tissue sections were cut from FFPE tissue blocks of host livers, skins, bone marrow biopsies, and graft liver biopsies from Renji Hospital (Fig. 1C). The tissue slides were deparaffinized in xylene (Adamas-beta, Shanghai, China) and rehydrated in a graded ethanol (Adamas-beta, Shanghai, China) series. Then, they were incubated in a preheated retrieval solution (R&D Systems, Boston, USA) at 95 °C for 30 min and cooled to room temperature. The tissue slides were then blocked with 3% BSA (Macklin, Shanghai, China) in DPBS (Gibco, San Francisco, USA) for 45 min at room temperature. After antigen retrieval, the tissue slides were incubated with an antibody cocktail, containing 19 metal isotope-tagged antibodies (Table S5) targeting immune cell markers and Tax, overnight at 4 °C in a humidified chamber. For nuclear staining, each section was stained for 30 min at room temperature with 100 µL of 1:600 diluted 191Ir/193Ir DNA intercalator (Fluidigm Sciences, San Francisco, USA) with DPBS.
Mass cytometry detection and subsequent analysis
After acquisition on Helios (Fluidigm, San Francisco, USA), data were normalized with four standard EQ beads (Fig. 1B). Mass cytometry data plots and histogram analysis were performed on CytoBank’s online platform (www.cytobank.org). A series of gates were used to select single cells and CD45+ cells, as depicted in Fig. S1. Parameters for dimensionality reduction were set, and PhenoGraph was applied to dimensionality reduction using the R package Cytofkit35. ArcSinh transformation, with a scaling factor of 5, was chosen as the transformation method to diminish the noise in these measurements.
IMC detection and subsequent analysis
Images were acquired with a Hyperion Laser Scanning Module (Fluidigm, San Francisco, USA) coupled to Helios mass cytometer (Fig. 1C). Fluidigm’s CyTOF v6.7 software-generated a.mcd file and a.txt file. The MCD Viewer v1.0 software was used for image processing and visualization.
ELISA essay
Serum samples were collected from the patients (ID:144 and ID:145) after diagnosing aGVHD. Blood samples were centrifuged for 5 minutes to separate serum. The serum was stored at -80°C. Samples were all tested by HTLV-I ELISA Kit (KNUDI, Quanzhou, China). All operations were carried out according to the kit instructions and the optical density was measured at 450 nm using a microtiter plate reader (BioTek, Winooski, USA).
Statistical analysis
Fisher’s exact test was adopted to test the differences in HTLV-1 positive rate between groups, and Benjamini-Hochberg adjustment was applied to control the false discovery rate. The adjusted p value <0.05 was considered statistically significant. In Fig. S3, the differences in HTLV-1 positive rate between groups were calculated by the Mann-Whitney test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets of the TargetSeq and CyTOF detections of the study are available on Zenodo (https://doi.org/10.5281/zenodo.7333323 and https://doi.org/10.5281/zenodo.7333412). The raw image files are available on Zenodo (https://doi.org/10.5281/zenodo.7333438). Source data are provided with this paper.
References
Paczesny, S., Hanauer, D., Sun, Y. & Reddy, P. New perspectives on the biology of acute GVHD. Bone Marrow Transplant. 45, 1–11 (2010).
Wysocki, C. A., Panoskaltsis-Mortari, A., Blazar, B. R. & Serody, J. S. Leukocyte migration and graft-versus-host disease. Blood 105, 4191–4199 (2005).
Kim, G. Y. et al. Dermatologic manifestations of solid organ transplantation-associated graft-versus-host disease: a systematic review. J. Am. Acad. Dermatol. 78, 1097 (2018).
Shulman, H. M. et al. Chronic graft-versus-host syndrome in man. A long-term clinicopathologic study of 20 Seattle patients. Am. J. Med. 69, 204–217 (1980).
Axt, L. et al. Retrospective single center analysis of outcome, risk factors and therapy in steroid refractory graft-versus-host disease after allogeneic hematopoietic cell transplantation. Bone Marrow Transplant. 54, 1805–1814 (2019).
Zhang, L., Yu, J. & Wei, W. Advance in targeted immunotherapy for graft-versus-host disease. Front. Immunol. 9, 1087 (2018).
Chen, X. B., Yang, J., Xu, M. Q., Wen, T. F. & Yan, L. N. Unsuccessful treatment of four patients with acute graft-vs-host disease after liver transplantation. World J. Gastroenterol. 18, 84–89 (2012).
Perri, R. et al. Graft vs. host disease after liver transplantation: a new approach is needed. Liver Transpl. 13, 1092–1099 (2007).
Dolgin, N. H. et al. Impact of recipient functional status on 1-year liver transplant outcomes. World J. Transplant. 9, 145–157 (2019).
Zeiser, R. & Blazar, B. R. Acute graft-versus-host disease - biologic process, prevention, and therapy. N. Engl. J. Med. 377, 2167–2179 (2017).
den Haan, J. M. et al. Identification of a graft versus host disease-associated human minor histocompatibility antigen. Science 268, 1476–1480 (1995).
Schlitt, H. J., Raddatz, G., Steinhoff, G., Wonigeit, K. & Pichlmayr, R. Passenger lymphocytes in human liver allografts and their potential role after transplantation. Transplantation 56, 951–955 (1993).
Hahn, A. B. & Baliga, P. Rapid method for the analysis of peripheral chimerism in suspected graft-versus-host disease after liver transplantation. Liver Transpl. 6, 180–184 (2000).
Manduzio, P. Transfusion-associated graft-versus-host disease: a concise review. Hematol. Rep. 10, 98–102 (2018).
Schroeder, M. L. Transfusion-associated graft-versus-host disease. Br. J. Haematol. 117, 275–287 (2002).
Petersdorf, E. W. The major histocompatibility complex: a model for understanding graft-versus-host disease. Blood 122, 1863–1872 (2013).
Matte-Martone, C., Liu, J., Jain, D., McNiff, J. & Shlomchik, W. D. CD8+ but not CD4+ T cells require cognate interactions with target tissues to mediate GVHD across only minor H antigens, whereas both CD4+ and CD8+ T cells require direct leukemic contact to mediate GVL. Blood 111, 3884–3892 (2008).
Kawase, T. et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood 110, 2235–2241 (2007).
Greene, W. C., Bohnlein, E. & Ballard, D. W. HIV-1, HTLV-1 and normal T-cell growth: transcriptional strategies and surprises. Immunol. Today 10, 272–278 (1989).
Coffin, J. M. The discovery of HTLV-1, the first pathogenic human retrovirus. Proc. Natl Acad. Sci. USA 112, 15525–15529 (2015).
Galli, V. et al. Essential role of human T cell leukemia virus type 1 orf-I in lethal proliferation of CD4(+) cells in humanized mice. J.Virol. 93, e00565-19 (2019).
Furuta, R. et al. Human T-cell leukemia virus type 1 infects multiple lineage hematopoietic cells in vivo. PLoS Pathog. 13, e1006722 (2017).
Ma, G. Y., Yasunaga, J. & Matsuoka, M. Multifaceted functions and roles of HBZ in HTLV-1 pathogenesis. Retrovirology 13, 100–110 (2016).
Moroso, V. et al. Donor and recipient HLA/KIR genotypes do not predict liver transplantation outcome. Transpl. Int. 24, 932–942 (2011).
Yagci, G. et al. The impact of donor variables on the outcome of orthotopic liver transplantation for hepatitis C. Transplant. Proc. 40, 219–223 (2008).
Zeiser, R. Advances in understanding the pathogenesis of graft-versus-host disease. Br. J. Haematol. 187, 563–572 (2019).
Fischer, J. C. et al. RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci. Transl. Med. 9, eaag2513 (2017).
Zeiser, R., Penack, O., Holler, E. & Idzko, M. Danger signals activating innate immunity in graft-versus-host disease. J. Mol. Med. 89, 833–845 (2011).
Fraser, C. et al. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 343, 1243727 (2014).
Bangham, C. R. M. Human T cell leukemia virus type 1: persistence and pathogenesis. Annu. Rev. Immunol. 36, 43–71 (2018).
Sobata, R. et al. Estimation of the infectious viral load required for transfusion-transmitted human T-lymphotropic virus type 1 infection (TT-HTLV-1) and of the effectiveness of leukocyte reduction in preventing TT-HTLV-1. Vox Sang. 109, 122–128 (2015).
Chang, Y. B. et al. Seroprevalence and demographic determinants of human T-lymphotropic virus type 1 and 2 infections among first-time blood donors-United States, 2000-2009. J. Infect. Dis. 209, 523–531 (2014).
Boxus, M. et al. The HTLV-1 tax interactome. Retrovirology 5, 76 (2008).
Brauweiler, A., Garrus, J. E., Reed, J. C. & Nyborg, J. K. Repression of Bax gene expression by the HTLV-I tax protein: Implications for suppression of apoptosis in virally infected cells. Virology 231, 135–140 (1997).
Chen, H. et al. Cytofkit: a bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput. Biol. 12, e1005112 (2016).
Iberg, C. A., Jones, A. & Hawiger, D. Dendritic cells as inducers of peripheral tolerance. Trends Immunol. 38, 793–804 (2017).
Iqbal, A. J. et al. Human CD68 promoter GFP transgenic mice allow analysis of monocyte to macrophage differentiation in vivo. Blood 124, e33–44 (2014).
Malavasi, F. et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol. Rev. 88, 841–886 (2008).
Ziegler, S. F., Ramsdell, F. & Alderson, M. R. The activation antigen CD69. Stem Cells 12, 456–465 (1994).
Kashem, S. W., Haniffa, M. & Kaplan, D. H. Antigen-presenting cells in the skin. Annu. Rev. Immunol. 35, 469–499 (2017).
Iliev, D. B., Thim, H., Lagos, L., Olsen, R. & Jorgensen, J. B. Homing of antigen-presenting cells in head kidney and spleen - salmon head kidney hosts diverse APC types. Front. Immunol. 4, 56–60 (2013).
Unanue, E. R. Antigen-presenting function of the macrophage. Annu Rev. Immunol. 2, 395–428 (1984).
Antohe, I. et al. The MHC-II antigen presentation machinery and B7 checkpoint ligands display distinctive patterns correlated with acute myeloid leukaemias blast cells HLA-DR expression. Immunobiology 226, 152049 (2021).
Wang, W. et al. Chromosome Y-encoded antigens associate with acute graft-versus-host disease in sex-mismatched stem cell transplant. Blood Adv. 2, 2419–2429 (2018).
Ali, S. A., Shah, V., McKinnon, R., Van Harn, M. & Janakiraman, N. Frequent expression of C4d in hepatic graft-versus-host disease: Potential clue for diagnosis and distinguishing acute and chronic form. Transpl. Immunol. 23, 77–80 (2010).
Ma, Q. et al. Complement component C3 mediates Th1/Th17 polarization in human T-cell activation and cutaneous GVHD. Bone Marrow Transplant. 49, 972–976 (2014).
Zhang, P. L., Wilkerson, M. L. & Schworer, C. M. C4d staining is a valuable marker in identifying chronic GVHD in colonic biopsies following BMT. Bone Marrow Transplant. 42, 209–211 (2008).
Saarloos, M. N., Koenig, R. E. & Spear, G. T. Elevated levels of iC3b and C4d, but not Bb, complement fragments from plasma of persons infected with human T cell leukemia virus (HTLV) with HTLV-I-associated myelopathy/tropical spastic paraparesis. J. Infect. Dis. 172, 1095–1097 (1995).
Kazama, T., Miyazawa, M., Tsuchiya, S. & Horii, A. Proliferation of macrophage-lineage cells in the bone marrow, severe thymic atrophy, and extramedullary hematopoiesis of possible donor origin in an autopsy case of post-transplantation graft-versus-host disease. Bone Marrow Transplant. 18, 437–441 (1996).
van Balen, P. et al. Tissue damage caused by myeloablative, but not non-myeloablative, conditioning before allogeneic stem cell transplantation results in dermal macrophage recruitment without active T-cell interaction. Front. Immunol. 9, 331 (2018).
Deguchi, M. et al. Comparison of the distribution and numbers of antigen-presenting cells among T-lymphocyte-mediated dermatoses: CD1a+, factor XIIIa+, and CD68+ cells in eczematous dermatitis, psoriasis, lichen planus and graft-versus-host disease. Arch. Dermatol. Res. 294, 297–302 (2002).
Rhodes, J. W. et al. Human anogenital monocyte-derived dendritic cells and langerin+cDC2 are major HIV target cells. Nat. Commun. 12, 2147 (2021).
Mair, F. & Liechti, T. Comprehensive phenotyping of human dendritic cells and monocytes. Cytom. Part A 99, 231–242 (2021).
Marino, J., Paster, J. & Benichou, G. Allorecognition by T lymphocytes and allograft rejection. Front. Immunol. 7, 94–100 (2016).
Lun, A. et al. Diagnostic value of peripheral blood T-cell activation and soluble IL-2 receptor for acute rejection in liver transplantation. Clin. Chim. Acta 320, 69–78 (2002).
Acknowledgements
We thank professor Chih-Ming Ho (University of California, Los Angeles) for his guidance, Dr. Q. Wei and Dr. J. Zheng (Department of pathology, Shanghai 10th People’s Hospital) for helping us with the immunohistochemistry, and professor Y. Ji (Zhongshan Hospital Affiliated to Fudan University) for performing the pathological diagnosis. We thank Dr. Wenqiong Su and Ms. Aiting Wang (Shanghai Jiao Tong University) for their technical assistance. This study was supported by the National natural science foundation of China (81871448). Fund beneficiary: X.D.
Author information
Authors and Affiliations
Contributions
Conceptualization, C.S. and Y.L.; Methodology, C.S. Y.L. B.W. and Q.X.; Investigation, Y.L. and C.S.; Formal Analysis, Y.L.; Writing – Original Draft, C.S. and Y.L.; Writing – Review & Editing, B.W., X.D., J.Z., Y.Q., Y.D., Z.Z. and N.M.; Visualization, Y.L.; Resources, C.S., T.L., and Z.Z.; Funding Acquisition, X.D., J.Z., and Q.X. Supervision, X.D. and Q.X.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Andrew Harman, Defu Zeng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shen, C., Li, Y., Wang, B. et al. HTLV-1 infection of donor-derived T cells might promote acute graft-versus-host disease following liver transplantation. Nat Commun 13, 7368 (2022). https://doi.org/10.1038/s41467-022-35111-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-35111-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.