Abstract
Osmotic stress imposed by drought and high salinity inhibits plant growth and crop yield. However, our current knowledge on the mechanism by which plants sense osmotic stress is still limited. Here, we identify the transcriptional regulator SEUSS (SEU) as a key player in hyperosmotic stress response in Arabidopsis. SEU rapidly coalesces into liquid-like nuclear condensates when extracellular osmolarity increases. The intrinsically disordered region 1 (IDR1) of SEU is responsible for its condensation. IDR1 undergoes conformational changes to adopt more compact states after an increase in molecular crowding both in vitro and in cells, and two predicted α-helical peptides are required. SEU condensation is indispensable for osmotic stress tolerance, and loss of SEU dramatically compromises the expression of stress tolerance genes. Our work uncovers a critical role of biomolecular condensates in cellular stress perception and response and expands our understanding of the osmotic stress pathway.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-sequencing datasets generated in this study can be found in the NCBI Gene Expression Omnibus under accession number GSE213934. The Arabidopsis genome information for mapping of RNA-sequencing reads is available from TAIR10 (www.arabidopsis.org). Source data are provided with this paper. All other data supporting the findings of this work are available within the paper and its Supplementary Information or from the corresponding authors on reasonable request.
References
Zhu, J. K. Abiotic stress signaling and responses in plants. Cell 167, 313–324 (2016).
Fabregas, N., Yoshida, T. & Fernie, A. R. Role of Raf-like kinases in SnRK2 activation and osmotic stress response in plants. Nat. Commun. 11, 6184 (2020).
Hoffmann, E. K., Lambert, I. H. & Pedersen, S. F. Physiology of cell volume regulation in vertebrates. Physiol. Rev. 89, 193–277 (2009).
Mourao, M. A., Hakim, J. B. & Schnell, S. Connecting the dots: the effects of macromolecular crowding on cell physiology. Biophys. J. 107, 2761–2766 (2014).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Oldfield, C. J. & Dunker, A. K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 83, 553–584 (2014).
Alberti, S. & Hyman, A. A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021).
Jung, J. H. et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256–260 (2020).
Chen, D. et al. Integration of light and temperature sensing by liquid-liquid phase separation of phytochrome B. Mol. Cell 82, 3015–3029 (2022).
Zhu, S. et al. Liquid–liquid phase separation of RBGD2/4 is required for heat stress resistance in Arabidopsis. Dev. Cell 57, 583–597 (2022).
Zhu, P., Lister, C. & Dean, C. Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression. Nature 599, 657–661 (2021).
Huang, X. et al. ROS regulated reversible protein phase separation synchronizes plant flowering. Nat. Chem. Biol. 17, 549–557 (2021).
Zavaliev, R., Mohan, R., Chen, T. & Dong, X. Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 182, 1093–1108 (2020).
Dorone, Y. et al. A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation. Cell 184, 4284–4298 (2021).
Kroschwald, S., Maharana, S. & Simon, A. Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters https://doi.org/10.19185/matters.201702000010 (2017).
Franks, R. G., Wang, C., Levin, J. Z. & Liu, Z. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129, 253–263 (2002).
Bao, F., Azhakanandam, S. & Franks, R. G. SEUSS and SEUSS-LIKE transcriptional adaptors regulate floral and embryonic development in Arabidopsis. Plant Physiol. 152, 821–836 (2010).
Huai, J. et al. SEUSS and PIF4 coordinately regulate light and temperature signaling pathways to control plant growth. Mol. Plant 11, 928–942 (2018).
Pfluger, J. & Zambryski, P. The role of SEUSS in auxin response and floral organ patterning. Development 131, 4697–4707 (2004).
Franks, R. G., Liu, Z. & Fischer, R. L. SEUSS and LEUNIG regulate cell proliferation, vascular development and organ polarity in Arabidopsis petals. Planta 224, 801–811 (2006).
Lee, J. E., Lampugnani, E. R., Bacic, A. & Golz, J. F. SEUSS and SEUSS-LIKE 2 coordinate auxin distribution and KNOXI activity during embryogenesis. Plant J. 80, 122–135 (2014).
Gong, X. et al. SEUSS integrates gibberellin signaling with transcriptional inputs from the SHR–SCR–SCL3 module to regulate middle cortex formation in the Arabidopsis root. Plant Physiol. 170, 1675–1683 (2016).
Zhai, H. et al. SEUSS integrates transcriptional and epigenetic control of root stem cell organizer specification. EMBO J. 39, e105047 (2020).
Zhang, X., Huai, J., Liu, S., Jin, J. & Lin, R. SIZ1-mediated SUMO modification of SEUSS regulates photomorphogenesis in Arabidopsis. Plant Commun. 1, 100080 (2020).
Rawat, P. et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell 81, 1013–1026 (2021).
Rana, P. S. et al. Calibration and characterization of intracellular asante potassium green probes, APG-2 and APG-4. Anal. Biochem. 567, 8–13 (2019).
Parker, J. C. Urea alters set point volume for K-Cl cotransport, Na-H exchange, and Ca-Na exchange in dog red blood cells. Am. J. Physiol. 265, C447–C452 (1993).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Boersma, A. J., Zuhorn, I. S. & Poolman, B. A sensor for quantification of macromolecular crowding in living cells. Nat. Methods 12, 227–229 (2015).
Cuevas-Velazquez, C. L. et al. Intrinsically disordered protein biosensor tracks the physical–chemical effects of osmotic stress on cells. Nat. Commun. 12, 5438 (2021).
Wallrabe, H. & Periasamy, A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16, 19–27 (2005).
Zhang, H., Zhao, Y. & Zhu, J. K. Thriving under stress: how plants balance growth and the stress response. Dev. Cell 55, 529–543 (2020).
Jing, Y. et al. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25, 242–256 (2013).
Sridhar, V. V., Surendrarao, A., Gonzalez, D., Conlan, R. S. & Liu, Z. Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc. Natl Acad. Sci. USA 101, 11494–11499 (2004).
Zhang, Q. et al. Visualizing dynamics of cell signaling in vivo with a phase separation-based kinase reporter. Mol. Cell 69, 334–346 (2018).
Tzamarias, D. & Struhl, K. Functional dissection of the yeast Cyc8–Tup1 transcriptional co-repressor complex. Nature 369, 758–761 (1994).
Oeser, M. L. et al. Dynamic sumoylation of a conserved transcription corepressor prevents persistent inclusion formation during hyperosmotic stress. PLoS Genet. 12, e1005809 (2016).
Nadel, C. M., Mackie, T. D. & Gardner, R. G. Osmolyte accumulation regulates the SUMOylation and inclusion dynamics of the prionogenic Cyc8–Tup1 transcription corepressor. PLoS Genet. 15, e1008115 (2019).
Liu, B. et al. Design and properties of genetically encoded probes for sensing macromolecular crowding. Biophys. J. 112, 1929–1939 (2017).
Liu, Z. & Karmarkar, V. Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13, 137–144 (2008).
Matsuyama, A. et al. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast 21, 1289–1305 (2004).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Gietz, R. D. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 1163, 33–44 (2014).
Rosa, S. Measuring dynamics of histone proteins by photobleaching in Arabidopsis roots. Methods Mol. Biol. 1675, 455–465 (2018).
Sandmann, M., Fuchs, J. & Lermontova, I. Immunolabeling of nuclei/chromosomes in Arabidopsis thaliana. Methods Mol. Biol. 1370, 127–135 (2016).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Acknowledgements
We thank X. Zhang for the synthesis of the b-isox compound. We thank L. Du for sharing the plasmids and yeast cells used in the heterologous expression system and Golz (The University of Melbourne) for sharing the seeds. This work was supported by grants from the National Natural Science Foundation of China to X.F. (32161133001), J.H. (31800235) and R.L. (32030009), Beijing Natural Science Foundation to X.F. (JQ21020) and the Strategic Priority Research Program of the Chinese Academy of Sciences to R.L. (XDB27030205).
Author information
Authors and Affiliations
Contributions
B.W., H.Z., J.H., R.L. and X.F. conceived the study. B.W. and J.W. performed the imaging of tobacco and Arabidopsis, RNA and genetic analyses. H.Z. performed b-isox precipitation imaging of yeast cells and all in vitro experiments. J.H. performed the phenotypic analyses. F.P. did all bioinformatic analyses. R.L. and X.F. wrote the manuscript. All authors contributed ideas and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 SEU undergoes condensation in response to hyperosmotic stress.
a, Illustration of the screen for osmotic stress-responsive biomolecular condensates. b, Representative confocal microscopic images of SEU-GFP expressed in yeast cells that were treated as indicated. Scale bars, 2 μm. c, Representative confocal microscopic images of tobacco epidermal cells expressing SEU-mVenus. The cells were treated as indicated. Scale bars, 5 μm. d, The protein levels of SEU and SEU-GFP in the indicated plants as determined by western blot. e, Expression of SEU relative to ACTIN in the indicated plants. Data are presented as the mean ± SD (n = 3 biologically independent experiments). Asterisk indicates a significant difference (two-tailed t test). f-i, Representative confocal microscopic images of Arabidopsis root tip cells expressing SEU-GFP. The roots were treated as indicated. Scale bars, 5 μm. j, Immunostaining of Arabidopsis root tip nuclei from plants treated with or without 300 mM NaCl. Scale bars, 5 μm. k, Representative confocal microscopic images of yeast cells expressing SEU. The cells were treated as indicated. Scale bars, 2 μm. l, Representative confocal microscopic images of tobacco epidermal cells expressing SEU. The cells were treated as indicated. Scale bars, 5 μm.
Extended Data Fig. 2 Analyses of SEU-Like proteins (SLKs).
a, Multiple sequence alignment of SEU and SLK protein sequences by Clustal W. b, The phylogenetic tree of SEU and SLK proteins inferred using the Neighbor-Joining method. c, The protein domain structures of SLKs. Disordered regions were predicted by IUpred algorithm. d, Representative confocal microscopic images of SLK2-GFP expressed in yeast cells that were treated by sorbitol for 30 min. Scale bars, 2 μm. e, The protein domain structure of a chimeric protein swapping the IDR1 of SLK2 with that of SEU. f, Representative confocal microscopic images of tobacco epidermal cells expressing the chimeric protein shown in (e). The cells were treated with or without NaCl for 5 min. Scale bars, 5 μm unless indicated.
Extended Data Fig. 3 SUMOylation of SEU is not required for its condensation in response to hyperosmotic stress.
a, The protein domain structure of SEU with four SUMOylation sites indicated. b, Representative confocal microscopic images of tobacco epidermal cells expressing SEU3KR-mVenus. The cells were treated with or without NaCl for 5 min. Scale bars, 5 μm. c-e, Confocal microscopy of Arabidopsis root tip cells of indicated genotypes. The roots were treated as indicated. Scale bars, 5 μm.
Extended Data Fig. 4 SEU condensation is sensitive to crowding.
a, Representative confocal microscopic images of yeast cells expressing IDR1-GFP. The cells were treated with or without NaCl and KCl for 5 min. Scale bars, 2 μm. b, Representative confocal microscopic images of yeast cells expressing SEU-IDR13KR-GFP. The cells were treated with or without sorbitol for 5 min. Scale bars, 2 μm. c, Representative confocal microscopic images of tobacco epidermal cells expressing SEU-mVenus under the ethanol inducible promoter. The cells were treated with 1% ethanol for indicated time. Scale bars, 5 μm. d, Coomassie staining of indicated protein samples before and after TEV cleavage to remove the MBP tag. e, In vitro phase separation assay of IDR1-mScarlet in the presence of indicated concentrations of PEG, NaCl or KCl. Scale bars, 5 μm.
Extended Data Fig. 5 IDR1 of SEU senses molecular crowding.
a, Illustration of IDR1 truncations. b, Representative confocal microscopic images of yeast cells expressing IDR1 truncations shown in (a) in the presence of 1.2 M sorbitol. Scale bars, 2 μm. c, In vitro phase separation assay of 0.2 μM IDR1-GFP protein in the presence of indicated concentrations of PEG8000. Scale bars, 5 μm. d, FRET with 0.2 μM IDR1 in the presence of indicated concentrations of PEG8000. e, Quantification of the donor fluorescence lifetime in yeast cells expressing indicated proteins. The cells were treated with 1.2 M sorbitol for 1 min. Asterisks indicate significant differences (****P ≤ 0.0001, two-tailed t test). 18 independent yeast cells were analyzed.
Extended Data Fig. 6 Phenotypic analysis of seu mutant.
a, The phenotypes of indicated genotypes grown horizontally on 1/2 MS media plates containing mannitol or NaCl. b, Photographs of indicated genotypes after 7 days of germination on 1/2 MS medium containing 0.5 μM ABA. c, Confocal microscopy of pSEU::SEU-GFP/seu transgenic root tip cells that were treated with 50 μM ABA for 20 min. Scale bars, 5 μm. d, Representative photographs of the indicated genotypes grown vertically on 1/2 MS media plates with or without 250 mM sorbitol under red light condition. e, The hypocotyl length of the indicated genotypes under sorbitol treatment relative to control. Data are presented as the mean ± SD (n ≥ 9 independent seedlings). Asterisks indicate significant differences (****P ≤ 0.0001, two-tailed t test).
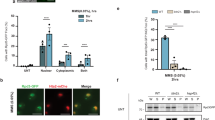
Extended Data Fig. 7 SEU is required for the expression of osmotic stress-tolerance genes.
a,b, Volcano plot of differentially expressed genes (DEGs) in seu-6 compared with Col-0 under control (a) or NaCl (b) treatment conditions determined by RNA‐sequencing. 10‐day‐old seedlings were transferred into liquid 1/2 MS medium or medium containing 300 mM NaCl and collected for RNA extraction after 15 min of treatment. Genes with at least 2-fold changes and P‐adjust less than 0.01 were considered as up- or down-regulated genes. c, Venn diagram showing the overlap of DEGs shown in (a) and (b). d, Heatmaps showing the relative expression changes (z‐normalized) of osmotic stress tolerance‐related genes under indicated conditions.
Extended Data Fig. 8 Condensation of SEU is evolutionarily conserved.
a, The phylogenetic tree of SEU homologs inferred using the Neighbor-Joining method. Evolutionary analysis was performed in MEGA X. b, Multiple sequence alignment of SEU homologs. The regions corresponding to IDR1, LDB and IDR2 were indicated. c, Zoom-in of the region marked by a rectangle in (b) showing the conservation of the two predicted α-helical peptides between SEU homologs. d, The structures of SEU homologs as predicted by AlphaFold. e, Representative confocal microscopic images of tobacco epidermal cells expressing SEU homologs. The cells were treated with or without NaCl for 5 min. Scale bars, 5 μm. f, The phylogenetic tree of AtSEU and its counterparts in yeast inferred using the Neighbor-Joining method. g, Top, protein domain structure of fission yeast Ssn6. Bottom, prediction of the disordered regions by IUpred algorithm. h, Representative confocal microscopic images of yeast cells expressing Ssn6. The cells were treated with or without sorbitol for 5 min. Scale bars, 2 μm.
Extended Data Fig. 9 A working model for SEU during osmotic response.
Under normal growth condition, SEU is diffused in the nucleus. When cells encounter hyperosmotic stress, shrinkage of cell volume leads to an increase of macromolecular crowding inside cells. Due to the hypersensitivity to crowding, IDR1 adopts more compact conformation, leading to the increase of local concentration and subsequent phase separation of SEU. Phase-separated SEU promotes the expression of stress-tolerance genes, thereby support cell survival.
Supplementary information
Supplementary Information
Supplementary Table 1.
Supplementary Data 1
List of DEGs in seu-6 mutants compared to wild-type Col-0.
Supplementary Video 1
Time-lapse imaging showing the recovery after half-bleaching of SEU nuclear condensates in tobacco epidermal cells. The red arrowhead indicates the area of bleaching. The photobleaching pulse was performed at time 3 s; scale bar, 2 μm.
Supplementary Video 2
Time-lapse imaging showing the condensation of IDR1SEU–GFP in yeast cells in the presence of 1.2 M sorbitol. Time 0 s indicates the addition of sorbitol; scale bars, 2 µm.
Supplementary Video 3
Time-lapse imaging showing the fusion of two IDR1–GFP droplets formed in vitro in the presence of 8% PEG8000; scale bar, 2 μm.
Supplementary Video 4
Time-lapse imaging showing the wetting of the slide surface by an IDR1–GFP droplet after contacting the slide; scale bar, 2 µm.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 4
Unprocessed western gels.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, B., Zhang, H., Huai, J. et al. Condensation of SEUSS promotes hyperosmotic stress tolerance in Arabidopsis. Nat Chem Biol 18, 1361–1369 (2022). https://doi.org/10.1038/s41589-022-01196-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01196-z
This article is cited by
-
Liquid–liquid phase separation of H3K27me3 reader BP1 regulates transcriptional repression
Genome Biology (2024)
-
The molecular basis for cellular function of intrinsically disordered protein regions
Nature Reviews Molecular Cell Biology (2024)
-
Systems engineering of Escherichia coli for high-level glutarate production from glucose
Nature Communications (2024)
-
A double-stranded RNA binding protein enhances drought resistance via protein phase separation in rice
Nature Communications (2024)
-
Technologies for studying phase-separated biomolecular condensates
Advanced Biotechnology (2024)