Abstract
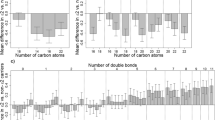
With the goal of identifying metabolites that significantly correlate with the protective e2 allele of the apolipoprotein E (APOE) gene, we established a consortium of five studies of healthy aging and extreme human longevity with 3545 participants. This consortium includes the New England Centenarian Study, the Baltimore Longitudinal Study of Aging, the Arivale study, the Longevity Genes Project/LonGenity studies, and the Long Life Family Study. We analyzed the association between APOE genotype groups E2 (e2e2 and e2e3 genotypes, N = 544), E3 (e3e3 genotypes, N = 2299), and E4 (e3e4 and e4e4 genotypes, N = 702) with metabolite profiles in the five studies and used fixed effect meta-analysis to aggregate the results. Our meta-analysis identified a signature of 19 metabolites that are significantly associated with the E2 genotype group at FDR < 10%. The group includes 10 glycerolipids and 4 glycerophospholipids that were all higher in E2 carriers compared to E3, with fold change ranging from 1.08 to 1.25. The organic acid 6-hydroxyindole sulfate, previously linked to changes in gut microbiome that were reflective of healthy aging and longevity, was also higher in E2 carriers compared to E3 carriers. Three sterol lipids and one sphingolipid species were significantly lower in carriers of the E2 genotype group. For some of these metabolites, the effect of the E2 genotype opposed the age effect. No metabolites reached a statistically significant association with the E4 group. This work confirms and expands previous results connecting the APOE gene to lipid regulation and suggests new links between the e2 allele, lipid metabolism, aging, and the gut-brain axis.

Similar content being viewed by others
References
Ji Y, et al. Apolipoprotein Epsilon epsilon4 frequency is increased among Chinese patients with frontotemporal dementia and Alzheimer’s disease. Dement Geriatr Cogn Disord. 2013;36(3–4):163–70.
Farrer LA, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis APOE and Alzheimer Disease Meta Analysis Consortium. Jama. 1997;278(16):1349–56.
Corbo RM, R Scacchi, Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ʻthriftyʼ allele? Ann Hum Genet, 1999; 63(Pt 4) 301–10
Sebastiani P, et al. APOE alleles and extreme human longevity. J Gerontol A Biol Sci Med Sci. 2019;74(1):44–51.
Sweigart B, et al. APOE E2/E2 is associated with slower rate of cognitive decline with age. J Alzheimers Dis, 2021.
Kim YJ, et al. Protective effects of APOE e2 against disease progression in subcortical vascular mild cognitive impairment patients: a three-year longitudinal study. Sci Rep. 2017;7(1):1910.
Reiman EM, et al. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat Commun. 2020;11(1):667.
Huang Y, Mahley RW. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiology of Disease. 2014;72:3–12.
Wang T, et al. APOE epsilon2 resilience for Alzheimer’s disease is mediated by plasma lipid species: analysis of three independent cohort studies. Alzheimers Dement, 2022.
Liu Y, et al. Plasma lipidome is dysregulated in Alzheimer’s disease and is associated with disease risk genes. Transl Psychiatry. 2021;11(1):344.
Hysi PG, et al. Metabolome genome-wide association study identifies 74 novel genomic regions influencing plasma metabolites levels. Metabolites. 2022;12(1):61.
Sebastiani P, Perls TT. The genetics of extreme longevity: lessons from the New England Centenarian Study. Front Genet. 2012;3:277.
Ferrucci L. The Baltimore Longitudinal Study of Aging (BLSA): a 50-year-long journey and plans for the future. J Gerontol A Biol Sci Med Sci. 2008;63(12):1416–9.
Earls JC, et al. Multi-omic biological age estimation and its correlation with wellness and disease phenotypes: a longitudinal study of 3,558 individuals. J Gerontol A Biol Sci Med Sci. 2019;74(Suppl_1):S52-s60.
Barzilai N, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290(15):2030–40.
Gubbi S, et al. Effect of exceptional parental longevity and lifestyle factors on prevalence of cardiovascular disease in offspring. Am J Cardiol. 2017;120(12):2170–5.
Wojczynski MK, et al. NIA Long Life Family Study: objectives, design, and heritability of cross sectional and longitudinal phenotypes. J Gerontol A Biol Sci Med Sci, 2021.
Wilmanski T, et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat Metab. 2021;3(2):274–86.
Sebastiani P, et al. A serum protein signature of APOE genotypes in centenarians. Aging Cell, 2019; e13023.
Fahy E, Subramaniam S. RefMet: a reference nomenclature for metabolomics. Nat Methods. 2020;17(12):1173–4.
Wishart DS, et al. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 2021;50(D1):D622–31.
Tuck MK, et al. Standard operating procedures for serum and plasma collection: Early Detection Research Network Consensus Statement Standard Operating Procedure Integration Working Group. J Proteome Res. 2009;8(1):113–7.
Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31(3):545–8.
Koch W et al. TaqMan systems for genotyping of disease-related polymorphisms present in the gene encoding apolipoprotein E. 2002;40(11):1123-1131.
Yamaguchi Y, et al. Plasma metabolites associated with chronic kidney disease and renal function in adults from the Baltimore Longitudinal Study of Aging. Metabolomics. 2021;17(1):9.
Mahieu NG, et al. Defining and detecting complex peak relationships in mass spectral data: the Mz unity Algorithm. Anal Chem. 2016;88(18):9037–46.
Cho K, et al. Targeting unique biological signals on the fly to improve MS/MS coverage and identification efficiency in metabolomics. Anal Chim Acta. 2021;1149:338210.
Stancliffe E, et al. DecoID improves identification rates in metabolomics through database-assisted MS/MS deconvolution. Nat Methods. 2021;18(7):779–87.
Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2006;8(1):118–27.
Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 2007;3(3):211–21.
Little, R.J.A. and D.B. Rubin, Statistical analysis with missing data. 1987: Wiley.
Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Statist Soc Series B. 1995;57(1):289–300.
Marron MM et al. Using lipid profiling to better characterize metabolic differences in apolipoprotein E (APOE) genotype among community-dwelling older Black men. Geroscience, 2021.
Proitsi P, et al. Plasma lipidomics analysis finds long chain cholesteryl esters to be associated with Alzheimer’s disease. Transl Psychiatry. 2015;5(1):e494–e494.
Mundra PA et al. Large-scale plasma lipidomic profiling identifies lipids that predict cardiovascular events in secondary prevention. JCI Insight; 2018. 3(17).
Baloni P et al. Multi-omic analyses characterize the ceramide/sphingomyelin pathway as a therapeutic target in Alzheimer’s disease. medRxiv, 2021 2021.07.16.21260601.
Wood PL, et al. Targeted lipidomics distinguishes patient subgroups in mild cognitive impairment (MCI) and late onset Alzheimer’s disease (LOAD). BBA clinical. 2015;5:25–8.
Wood PL, et al. Targeted lipidomics of fontal cortex and plasma diacylglycerols (DAG) in mild cognitive impairment and Alzheimer’s disease: validation of DAG accumulation early in the pathophysiology of Alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2015;48(2):537–46.
Wood PL, Cebak JE, Woltjer RL. Diacylglycerols as biomarkers of sustained immune activation in proteinopathies associated with dementia. Clinica Chimica Acta. 2018;476:107–10.
Callender JA, Newton AC. Conventional protein kinase C in the brain: 40 years later. Neuronal signaling. 2017;1(2):NS0160005-NS20160005.
Soto-Avellaneda A, Morrison BE. Signaling and other functions of lipids in autophagy: a review. Lipids Health Dis. 2020;19(1):214.
Herrera JJ et al. Acarbose has sex-dependent and -independent effects on age-related physical function, cardiac health, and lipid biology. JCI Insight, 2020; 5(21).
Sonowal R, et al. Indoles from commensal bacteria extend healthspan. Proc Natl Acad Sci. 2017;114(36):E7506–15.
Masuo Y, et al. 6-Hydroxyindole is an endogenous long-lasting OATP1B1 inhibitor elevated in renal failure patients. Drug Metab Pharmacokinet. 2020;35(6):555–62.
Dekkers KF et al. An online atlas of human plasma metabolite signatures of gut microbiome composition. medRxiv, 2021 2021.12.23.21268179.
Badal VD, et al. The gut microbiome, aging, and longevity: a systematic review. Nutrients. 2020;12(12):3759.
Zajac DJ, et al. APOE genetics influence murine gut microbiome. Sci Rep. 2022;12(1):1906.
Crumeyrolle-Arias M, et al. Inhibition of brain mitochondrial monoamine oxidases by the endogenous compound 5-hydroxyoxindole. Biochem Pharmacol. 2004;67(5):977–9.
Behl T et al. Role of monoamine oxidase activity in Alzheimer’s disease: an insight into the therapeutic potential of inhibitors. Molecules, 2021; 26(12).
Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10(5):333–44.
Wong MWK, et al. APOE genotype differentially modulates plasma lipids in healthy older individuals, with relevance to brain health. J Alzheimers Dis. 2019;72(3):703–16.
Miranda AM, et al. Effects of APOE4 allelic dosage on lipidomic signatures in the entorhinal cortex of aged mice. Transl Psychiatry. 2022;12(1):129.
Gurinovich A et al. Effect of longevity genetic variants on the molecular aging rate. Geroscience, 2021.
Acknowledgements
BLSA was supported by the Intramural Research Program of the National Institute on Aging. We acknowledge Mr. Brendan A. Mitchell for statistical assistance.
Funding
NIA R01AG061844 (PS, TTP), NIA U19-AG023122 (to NR, TTP, PS); USDA 58–1950-4–003 (MSL), NHLBI T32-HL083825 (MMM). NIA R01AG057909 (NB, SM), NIA R01AG061155 (SM, NB). NIA U19AG063893 (GJP, TTP), NIA UH2AG064704 (TTP, PS, SM, NB). This project was supported in part by the USDA Agricultural Research Service Cooperative Agreement 58-8050-9-004. The content is the sole responsibility of the authors and does not necessarily represent the official views of the USDA.
The authors declare no competing interests. All study participants provided informed consent and the studies were approved by the Institutions’ IRB as described in the “Methods.”
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Sebastiani, P., Song, Z., Ellis, D. et al. A metabolomic signature of the APOE2 allele. GeroScience 45, 415–426 (2023). https://doi.org/10.1007/s11357-022-00646-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-022-00646-9