Discovery of a hidden form of neuropeptide F and its presence throughout the CNS–gut axis in the mud crab, Scylla olivacea
- 1Department of Anatomy, Faculty of Science, Mahidol University, Bangkok, Thailand
- 2Center of Excellence for Shrimp Molecular Biology and Biotechnology (CENTEX Shrimp), Faculty of Science, Mahidol University, Bangkok, Thailand
- 3Chulabhorn International College of Medicine, Thammasat University, Pathumthani, Thailand
- 4Academic and Curriculum Division, Nakhonsawan Campus, Mahidol University, Nakhonsawan, Thailand
The mud crab Scylla olivacea (Scyol) is an economically crucial crustacean species in Thailand, due to its high market demand and nutritional value. The neuropeptide F (NPF) has been implicated in the coordinated regulation of feeding and metabolism in invertebrates. While various isoforms of neuropeptide F (NPF) have been previously explored in the mud crab, some knowledge gaps in relation to the NPF family, and ambiguities in the nomenclature from previous reports, remain. In this study, NPF was firstly localized in the central nervous system and gastrointestinal tract of the mud crab, S. olivacea, using a polyclonal antibody against Macrobrachium rosenbergii, Macro-NPF. The NPF immunoreactivity (ir) was detected dominantly in the X-organ/sinus gland complex of the eyestalk (ES) and the various neuronal clusters (cluster 6, 9/11, and 14/15) and neuropils (anteromedial and posteromedial protocerebral neuropils, olfactory and accessory olfactory neuropils, and medial antennule neuropil, columnar neuropil) of the brain (BR), commissural ganglia, and suboesophageal ganglion of the ventral nerve cord (VNC). Interestingly, this study also presented the NPF immunoreactivity (NPF-ir) in the acinar gland-like cell and spindle-shaped epithelial cells of S. olivacea intestine. The full-length Scyol-NPFII was characterized by molecular cloning and revealed 414 nucleotides with 375 nucleotides of an open reading frame which encoded 124 deduced amino acids. A 124-amino acid precursor protein of Scyol-NPFII included a 26-residue signal peptide and a 69-amino acid mature peptide. The Scyol-NPFII showed the highest percentage of hit similarity to S. paramamosain-NPFII and clustered in the NPFII family, separated from the other forms of NPFs in this species. The spatial gene expression in various tissues revealed that Scyol-NPFII was found dominantly in the ES and BR, VNC, heart, intestine, and muscle. This study provided a novel form of NPF in the female mud crab, S. olivacea, which could open the possibility of its functioning in the brain-to-gut controlling axis. This study could provide essential information for further application in the cultured system of S. olivacea in the near future.
Introduction
The current status of mud crab (Scylla olivacea) hatchery production in Thailand is unpredictable to increasing market demands due to the small scale of harvesting and culturing. Currently, the grow-out and fattening of lean crabs after harvesting from natural sources is the most common practice in the Scylla spp. culture system (Nooseng, 2015). For the sustainable breeding culture of this species, the monitoring of growth and reproductive cycling of broodstock is essential. Massive muscle growth after growing in pond culture would be expected with intensive strategies, including culture with a short duration of feeding, gaining of high growth performance by the requirement of low feeding, and the cross-species hybridizing for new-offspring production (Shelley, 2008; Cui et al., 2021). However, all strategies above remain facing the abnormality in morphological characteristics of the hybrid offspring crab due to genetic problems in exoskeletal formation via the chitin biosynthetic pathway (Farhadi et al., 2022) and may require the internal factors via the endocrine system, which may regulate the physiological functions in the species (Duangprom et al., 2017; Kornthong et al., 2019).
Neuropeptide F (NPF) is orthologous in function with vertebrate neuropeptide Y (NPY) (Fadda et al., 2019). It is well known that NPY plays an essential role in feeding regulation in the “brain–gut axis”, facilitating appetite and obesity regulation in humans (Wu et al., 2019). In invertebrates, the NPF plays various roles in diverse functions, expanding throughout several biological responses, including feeding-metabolic regulation, reproductive and sexual responses, ethanol sensing, locomotor circadian rhythms, and learning-stress responses (Cui and Zhao, 2020). NPF is generally referred to as a peptide that consists of more than 28 amino acids and contains the “RXRF-amide” carboxyterminal motif (Fadda et al., 2019). However, another form of neuropeptide F is the short neuropeptide F (sNPF) which consists of eight to 12 residues and typically contains the “M/T/L/FRF-amide” carboxyterminal motif (Nässel and Wegener, 2011; Fadda et al., 2019). However, it remained facing the ambiguities of the NPF nomenclature that was used in many previous studies. Moreover, NPF could be suggested to play the overlapping roles with sNPF in feeding and metabolism controls in invertebrates, but taken place with different mechanisms (Fadda et al., 2019). In the arthropods, NPF was identified in the suboesophageal ganglion and midgut of the yellow fever mosquito, Aedes aegypti, based on immunohistochemistry, playing a role in inhibiting transepithelial ion transportation (Onken et al., 2004). In Drosophila, the npf transcript is expressed in a pair of neurons in the suboesophageal ganglion, which showed the role in sugar sensing and ingestion at the larval stage (Shen and Cai, 2001). However, NPF’s expression pattern in Drosophila sp. fluctuated, according to the stage-specific feeding phenotypes (Wu et al., 2003). Recently, the enteroendocrine cell (EEC) in the midgut of D. melanogaster produced and released NPF, which was proved to act as an incretin-like hormone. This evidence also suggested that the enteroendocrine NPF affected metabolic function through glucagon-like and insulin-like hormones (Yoshinari et al., 2021). Taken together, NPF was the essential neuropeptide that played a major role in the CNS–gut axis in insects like a presence in humans (Yoshinari et al., 2021).
A range of studies have reported NPF in many species of crustaceans, including Marsupenaeus vannamei, Daphnia magna (Christie et al., 2008), Litopenaeus vannamei, Melicertus marginatus (Christie et al., 2011), and Macrobrachium rosenbergii (Suwansa-Ard et al., 2015; Thongrod et al., 2017). In S. olivacea, an NPF and two isoforms of sNPF were predicted by the publicly accessible transcriptome shotgun assembly technique (Christie, 2016). Comparatively, Scylla paramamosain NPFs were reported earlier, including two transcripts of NPFs which encoded complete RPRFamide peptide precursors and one sNPF that encoded the XPXRLRFamide-conserved motif (Bao et al., 2015). All of them showed the pattern of expression throughout the CNS, whereas sNPF was dominantly expressed in the ovary, suggesting that it may play a role during the vitellogenesis of this species.
Localization and expression patterns of S. olivacea NPF remain uninvestigated, and some forms of NPF in this species remain undiscovered. Therefore, this study primarily aimed to demonstrate NPF mapping in the central nervous system and alimentary tract of this species, proposing a brain-gut axis, using immunohistochemistry with a fluorescent-based technique, probing with a specific M. rosenbergii NPF I (Macro-NPF I) antibody (Thongrod et al., 2017). We hypothesized that there is a hidden NPF isoform that has never been found in S. olivacea. Hence, the NPF-II isoform’s characteristics in this species, corresponding to the specific binding with the Macro-NPF I antibody, was also investigated by molecular cloning, multiple alignments, and phylogeny analysis, compared to the NPF member of other crustaceans. In addition, the reactivity of the Macro-NPF I antibody to S. olivacea tissue was proved by dot-blot analysis and immunohistochemistry.
Materials and methods
Experimental animals and animal ethic statement
Healthy mature female S. olivacea were obtained from local farms in Ranong province, Thailand. The maturing status of the crabs was determined using the previously described methods (Ikhwanuddin et al., 2011). Based on size, mature female mud crabs were 9 to 12 cm in carapace width and 150 to 300 g in body weight. Their ovarian stage was examined and classified, based on morphological appearance (Islam et al., 2010). The crabs were maintained in concrete tanks following the previous studies (Duangprom et al., 2017; Duangprom et al., 2018). The animal use for tissue collection was performed according to the guidelines on the care and use of animals for scientific purposes provided by the Institutional Care and Use Committee (ICCUC) of Thammasat University, which approved this specific study in the National Research Council of Thailand (NRCT) (Protocol no. 011/2562). ICCUC approved the tissue sample use through the Faculty of Science, Mahidol University’s statement (Protocol no. MUSC64-025-574). All efforts were made to minimize animal suffering.
Tissue collection for immunohistochemistry
The central nervous tissues (CNS), eyestalk (ES), brain (BR), ventral nerve cord (VNC), and the alimentary tissues, including stomach (ST), intestine (IN), and hepatopancreas (HP), were collected from the animals after euthanasia in a cooling ice basket. The neural and non-neural tissues were immersed into 4% paraformaldehyde fixative for overnight, at a chill temperature (4°C). The fixed tissues were washed with chill PBS several times to remove the residual fixative and kept in PBS containing 0.1% sodium azide for a few weeks before being subsequently used.
Immunofluorescence, whole-mount immunofluorescence, and immunohistochemistry
The protocol of wholemount immunofluorescence followed Kruangkum et al. (2013) and Thongrod et al. (2017). Briefly, fixed whole-CNS tissues were cleaned and de-sheathed under a stereomicroscope (Nikon, SMZ745T). They were immersed in 0.02% PBS with Triton X-100 (PBST) for a day at 4°C. To increase the permeability of the tissues, they were transferred into a solution containing 80% methanol and 20% DMSO at -20°C for 5–6 h. The tissues were washed in PBS and PBST several times before being incubated with the primary antibody solution containing a 1:500 concentration of rabbit anti-NPF antibody (anti-Macro-NPF I) (special gift from Thongrod et al., 2017) in blocking solution, containing 10% normal goat serum in PBST. After being immersed in a solution containing antibody for 4 days at 4°C, the tissues were washed several times in PBS and PBST, for at least 10 min each step. They were finally incubated in a solution containing a secondary antibody (1:1,000 Alexa 488 (goat anti-rabbit IgG Alexa Fluor® 488, ab150077, Lot: GR3181607-3, Abcam, USA) in blocking solution) for 3 days. The nuclear localization was stained with the molecular nuclear probes (Hoechst 33342 (Molecular Probes, Eugene, OR, USA), DAPI (Invitrogen™), or TOPRO3 (Invitrogen™)) by mixing in a solution of secondary antibody. After washing, all tissues were dehydrated with increasing gradient concentrations of ethanol before clearing the tissues with methylsalycylate. The sample observation and photography were analyzed by the confocal microscope (Leica TCS SP8) with LAS-X Software and 3D Visualization Software Module.
Immunofluorescence and immunohistochemistry were subsequently performed on alimentary organ tissue of 5–10-μm thickness, sectioned by vibratome or microtome. The tissue materials were blocked by non-specific binding with 1% glycine in PBS before incubating with a 10% blocking solution for an hour. The primary antibody with 1:500 dilution was applied overnight at 4°C. The tissues were then incubated in secondary antibodies [(goat anti-rabbit IgG Alexa Fluor® 488, ab150077, Lot: GR3181607-3, Abcam, USA) and (goat anti-mouse IgG Alexa Fluor® 647 Lot: GR3212989-1, Abcam, USA)] with TOPRO3 in blocking solution for immunofluorescent technique and goat anti-rabbit IgG HPR (goat anti-rabbit IgG-HRP, sc-2004, Lot # F2612, Santa Cruz Biotechnology) counterstaining with hematoxylin nuclear staining for HRP-immunohistochemistry, whereas the negative control sections were applied by pre-immune serum of primary antibody. All tissue sections were observed by confocal microscopy (Olympus FV1000) for immunofluorescence detection and light microscopy (Leica DM750) with a photography detector (Leica ICC50 HD) for immunohistochemistry.
RNA isolation
The nervous tissues and non-nervous tissues, including muscle (MU), gill (GI), and heart (HT), and the alimentary tissues were collected from female S. olivacea and immediately frozen in liquid nitrogen before being stored at -80°C until use. Total RNA was extracted from each tissue using TriPure Isolation Reagent (Roche, Germany), following the manufacturer’s protocol. The purity and quantity of each RNA sample were measured by a NanoDrop spectrophotometer at 260 and 280 nm.
Gene validation and molecular cloning
Total RNA (2 μg) extracted from the CNS of female S. olivacea was used for first-strand cDNA synthesis using QuantiNova Reverse Transcription Kit (Qiagen, Germany), following the manufacturer’s protocol. For polymerase chain reaction (PCR), there was amplification of the Scyol-NPF II transcript, and primers (Table 1) were designed from an NPF transcript derived from S. paramamosain and M. rosenbergii (Bao et al., 2015; Thongrod et al., 2017). Complementary DNA was subsequently used as a template for PCR using forward and reverse primers with details included in Table 1. The thermocycling condition used for PCR amplification was set as follows: one cycle at 94°C for 5 min, followed by 35 cycles of 30 s at 94°C, 45 s at 55°C, and 45 s at 72°C, with a final extension of 10 min at 72°C. The PCR product was further analyzed by gel electrophoresis using 2% agarose gel. The amplicon was purified using a GeneJET Gel Extraction Kit (Thermo Scientific, USA) and cloned into a pGEM® T Easy Vector (Promega, USA). Plasmids with insert sequences were purified using a GeneJET Plasmid Miniprep Kit (Thermo Scientific, USA), then sequenced by Macrogen (Macrogen Inc., Korea). The sequence identification was conducted according to the previously described protocol (Duangprom et al., 2018; Kornthong et al., 2019).
Transcript distribution by reverse transcription-polymerase chain reaction
The cDNA from various tissues, as mentioned above, were used as templates for reverse transcription-polymerase chain reaction (RT-PCR). The condition of PCR was performed according to the method previously described by Duangprom et al. (2018) and Kornthong et al. (2019). To investigate Scyol-NPF II expression, gene-specific primers (NPF118-F and NPF443-R) were used (Table 1). The RT-PCR exponential phase was determined within 30 cycles to allow the comparison of cDNAs developed from an internal control (Scyol-β-actin).
Sequence analysis and phylogeny
The full-length Scyol-NPF II nucleotide sequence was used for Basic Local Alignment Search Tool (BLAST) search against the National Center for Biotechnology Information (NCBI) GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for defining similar sequences. The Scyol-NPF II nucleotide sequence was predicted and translated to an amino acid sequence using the Expasy bioinformatics resource portal-translate tool (http://web.expasy.org/translate/). Protein sequences of NPF from other related species were retrieved from Entrez (NCBI). BLAST hit transcripts were retrieved, translated into amino acid sequences, and subsequently used in multiple-sequence alignments for the comparison. Multiple-sequence alignment among the NPF family and NPF-II of several species was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and presented with BOXSHADE software (http://sourceforge.net/projects/boxshade/). The amino acid sequence logos were generated using the WebLogo tool (Crooks et al., 2004). Phylogenetic tree analysis was conducted using MEGA X software (Stecher et al., 2020) based on the Neighbor-Joining method with a bootstrap consensus tree.
A pre-absorption antibody with Scyol-NPF II synthetic peptide and dot-blot analysis
Based on the peptide sequence from this study, the Scyol-NPF II peptide with the sequence “KPDPTQLAQMADVIKYLHELDKYYSPMSRPSPRSAPGPASQIQALENTLKFLQLQELGKKYSHVTRPRFamide” was commercially synthesized by ProteoGenix (Schiltigheim, France). Macro-NPF I antibody used in the experiment was kindly provided and supported by Dr. Sirorat Thongrod (Thongrod et al., 2017). The antibody specificity and cross-reactivity of anti-Macro-NPF I for the Scyol-NPF II synthetic peptide were tested by dot-blot analysis. The diluted synthetic Scyol-NPF II in distilled water at 1-mg/ml concentration was shaken-incubated with 1 ml of Macro-NPF antibody overnight at 4°C, as a preabsorbed antibody. It can be stored at -20°C for several weeks before use. The diluted synthetic peptide at 1- and 0.5-mg/ml concentrations and a 500-ng/μl concentration of BSA were dotted onto a 0.45-mm nitrocellulose membrane and then dried for a few hours before applying the blocking solution, which contained 5% BSA in PBST onto the membrane for 2 h at room temperature. Then, the primary antibody was used in two separated conditions in the dot-blot analysis: (1) preabsorbed-Scyol-NPF II peptide with anti-Macro-NPF I antibody and (2) anti-Macro-NPF I antibody, diluted in PBST overnight at 4°C. They were gently shaken throughout the experiment including several washing steps of 10 min each. Then, the secondary antibody solution containing HRP-conjugated goat anti-rabbit IgG H&L (Abcam, ab6721, USA) was used at a 1:5,000 dilution in PBS for 1 h. Finally, an enhanced chemiluminescence kit (Thermo Fisher Scientific Inc., Pittsburgh, PA) was used to develop the membranes and the result was detected and photographed using the Amersham Imager (GE Healthcare, Sweden).
Results
NPF distribution and mapping in the CNS
The localization of NPF immunoreactivity (NPF-ir) was observed throughout the central nervous tissues (CNS) including eyestalk (ES), brain (BR), and ventral nerve cord (VNC) of the mud crab, S. olivacea. Wholemount immunofluorescence revealed the overall mapping distribution of NPF-ir which was revealed throughout in neuronal clusters, neuropils, and nerve fibers which communicated inside and connected to the brain via the protocerebral tract (PT) (Figure 1A), especially in the X-organ-sinus gland complex (XO-SG) of ES, including their connecting fibers (Figures 1B, C). Moreover, ES-associated neuropils, including the medulla externa, medulla interna, and medulla terminalis, were also scattered with intense immunoreactive signals (Figures 1A, D). The small neurons in clusters 2 and 3 (Figure 1A) and medium-sized neurons in clusters 3–4 were also revealed (Figure 1E).
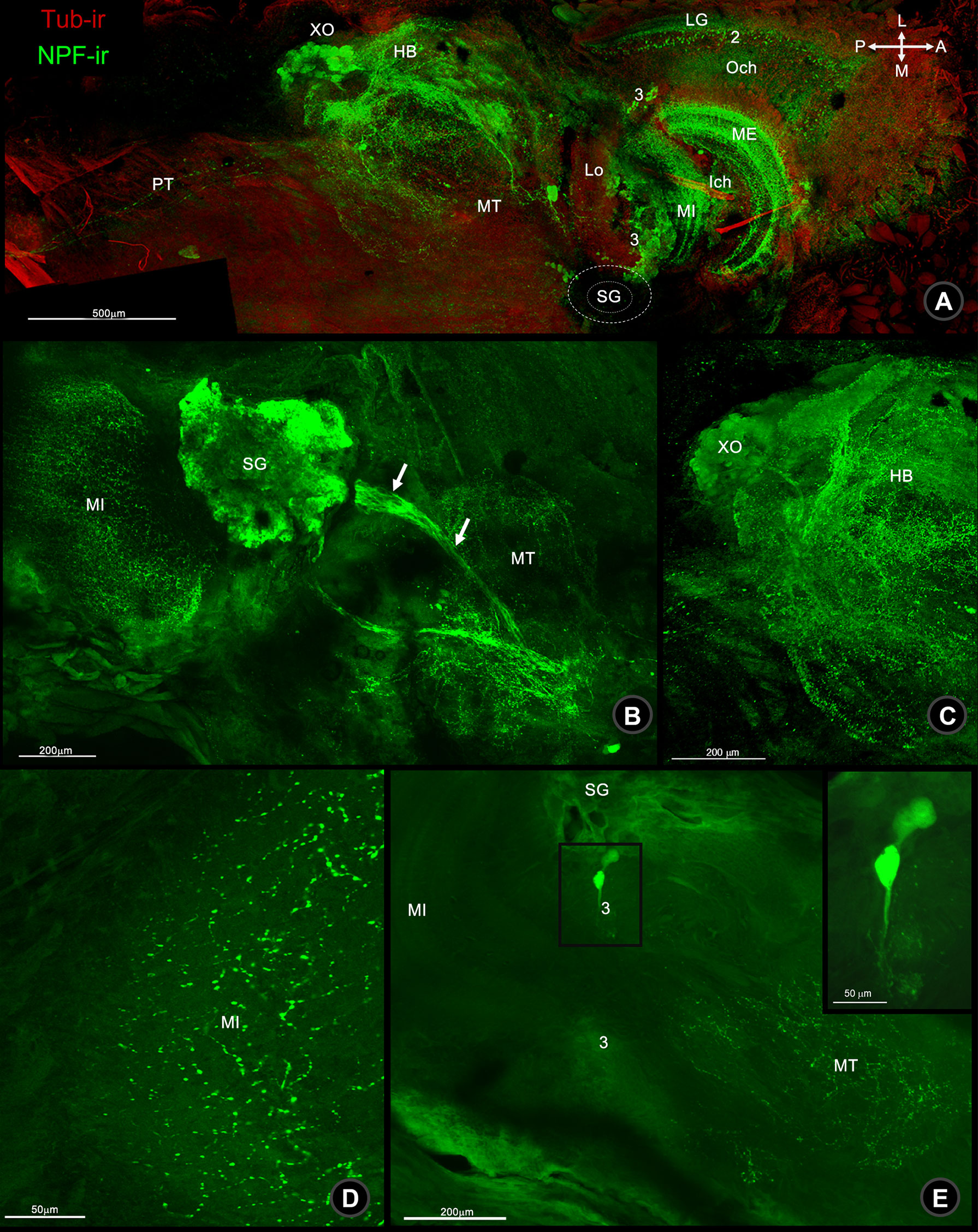
Figure 1 Wholemount immunofluorescence photographs of Macro-NPF-ir in the bi-section of the mud crab’s ES. (A) Low-magnification overview image of ES revealed the localization of Macro-NPF-ir in the neuropils and neuronal clusters. NPF-ir showed in SG and its connecting fibers to XO (arrows) (B) the fiber network of HB and XO (C), nerve fiber punctuates forming as medullary interna (MI) (D), and medium neurons in cluster 3 (E). (LG, laminar ganglionaris; Och, outer optic chiasm; ME, medullar externa; Ich, inner optic chiasm; MI, medullar interna; Lo, lobar; SG, sinus gland; MT, medullar terminalis; HB, hemiellipsoid body; XO, X-organ; PT, protocerebral tract).
In the brain (BR), NPF-ir was revealed in many parts, including in the protocerebrum (PC), deutocerebrum (DC), and tritocerebrum (TC) (Figure 2A). Strong signals were observed in the neuropil of the protocerebrum, including the anteromedial protocerebral neuropil (AMPN), posteromedial protocerebral neuropil (PMPN), and PT, which links the brain and ES (Figures 2A–C). However, a few neurons in cluster 6 were also observed in NPF-ir (Figure 2C). In DC, a pair of olfactory neuropil (ON), accessory lobe of olfactory neuropil (AcN), and median antenna neuropil (MAN), as well as sub-cap regions and internal punctate were detected NPF-ir (Figures 2A, D and Supplement 1A). In contrast, NPF-ir was observed intensely in the medium-sized neurons of cluster 9/11, while there was mild staining in small neurons (Figures 2D, Supplement 1A, 2). In TC, strong-intensity NPF-ir was observed in a pair of the antenna neuropil (AnN) and columnar neuropil (CN), and large-size neurons in cluster 14/15 (Figure 2D).
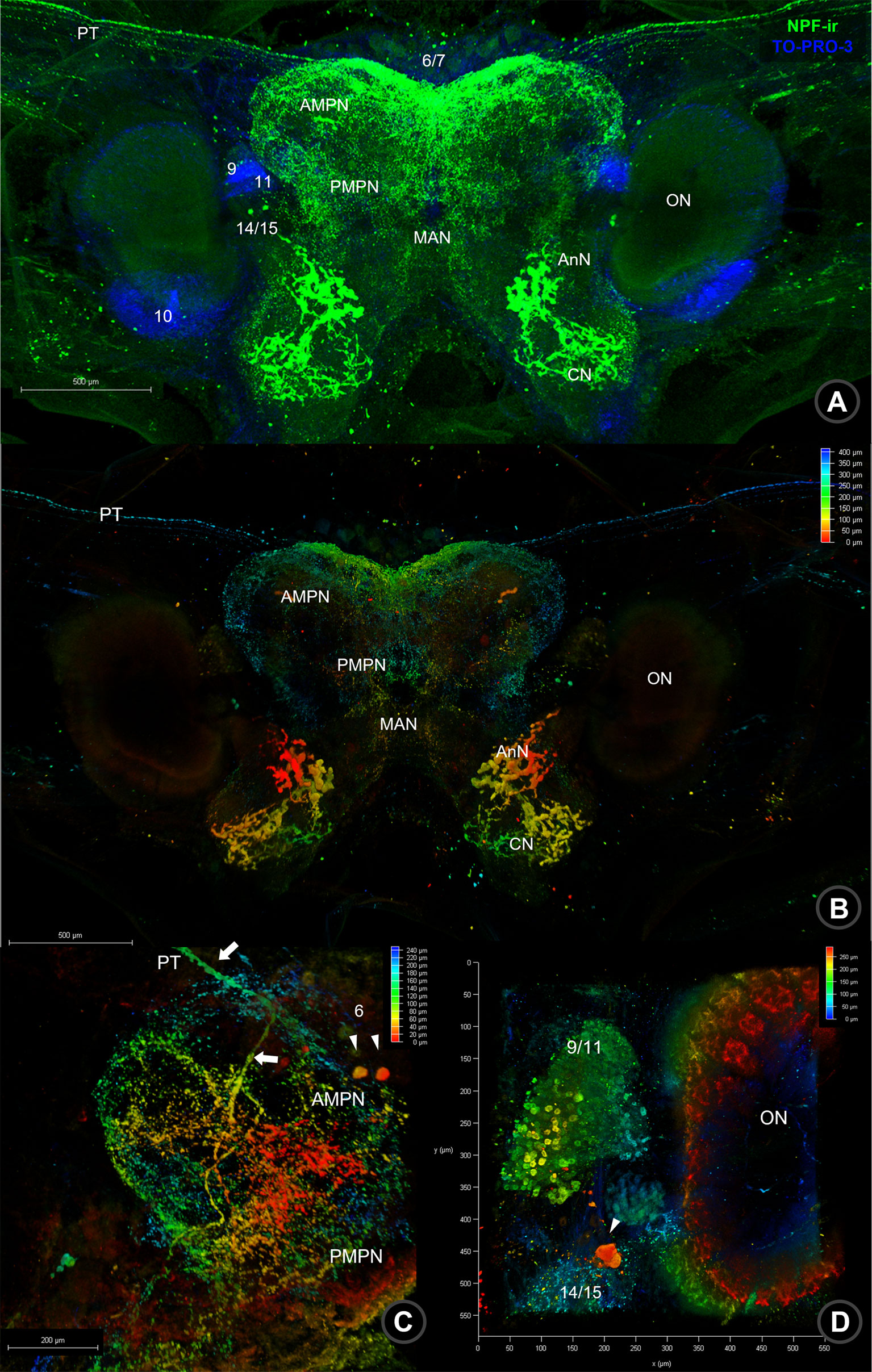
Figure 2 Wholemount immunofluorescence photographs of Macro-NPF-ir in the brain. (A) The fluorescence detection of NPF-ir was detected in the whole brain; nuclear staining is presented by TOPRO-3. (B) Depth code photographs of NPF-ir in the whole brain indicated the different plane of localization in neuropils and neurons. (C) Depth code photograph of NPF-ir localized in neuroplis, including AMPN, PMPN, nerve fiber (PT) connecting to the ES, and neurons in cluster 6. (D) NPF-ir presented in the ON and neurons in cluster 9/11 of the deutocerebrum and clusters 14–15 of the tritocerebrum. The depth coding represented thickness and NPF-ir signal localizing in different layers from the point of indication. (PT, protocerebral tract; AMPN, anteriomedial protocerebral neuropil; PMPN, poteriomedial protocerebral neuropil; MAN, medial antenna I neuropil; AnN, antenna I neuropil; CN, columnar neuropil).
In the last part of the CNS, the ventral nerve cord (VNC), the signal of NPF-ir was detected continuously in the circumoesophageal connective (CeC) fiber, linking fibers between BR and VNC. A positive signal of NPF was observed in the commissural ganglia (CoG), which contain small and medium-sized neurons and their neuropils (Figure 3A). In the suboesophageal ganglion (SEG), NPF-ir was dominantly observed in the visceral sensory neuropil (VSN), maxillary neuropils (MN), and the related neurons, especially a pair of large neurons (Figure 3B) as well as the neurons in the dorsolateral cluster (DLC) (see Supplement 1B, 3). Moreover, the signals were also observed in abundant small and medium-sized neurons and the punctate/neuropils of SEG (Figure 3C), thoracic neuropils (Figure 3D), and abdominal neuropils (Figures 3E–G and Supplement 4).
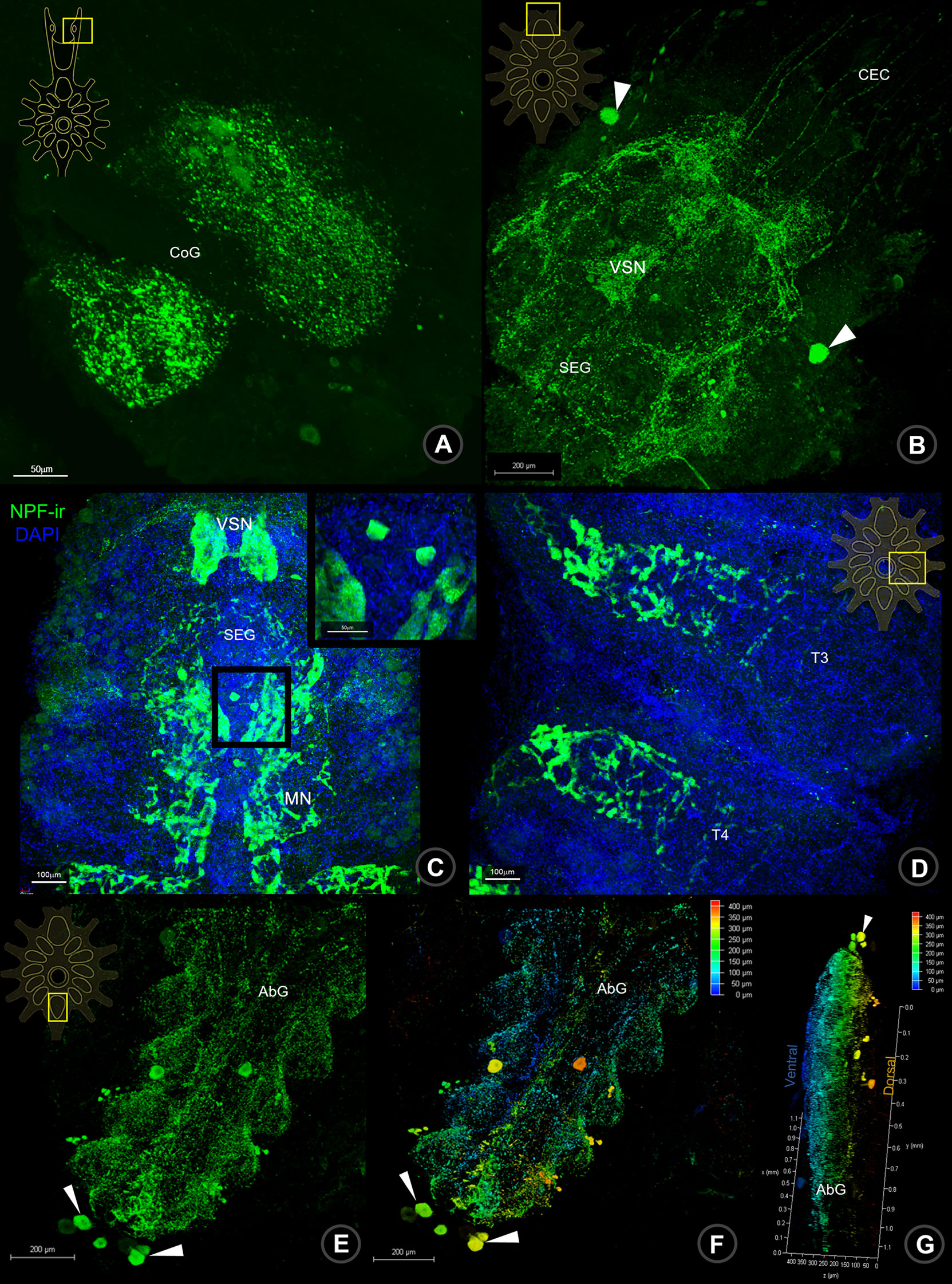
Figure 3 Wholemount immunofluorescence photographs of Macro-NPF-ir in the ventral nerve cord. The NPF-ir detection presented in CoG (A), nerve fibers of CEC, neurons (arrows) and neuropil of SEG (B, C), thoracic neuropils (D), neurons (arrows) and neuropils of AbG (E). The ventral (F) and lateral views (G) of depth coding photographs of NPF-ir shown in AbG. CoG, commissural ganglion; CEC, circumesophageal connection; SEG, subesophageal ganglion; T, thoracic ganglion; AbG, abdominal ganglion.
NPF immunolabeling in the gastrointestinal tract
S. olivacea intestinal structure identification was based on the previously described structure in Procambarus clarkii (To et al., 2004). NPF-ir was revealed in the intestinal structures of S. olivacea by whole-mount immunofluorescence and immunohistochemistry. The localization pattern of NPF-ir was found in the longitudinal nerve fibers that innervated in the outermost serosa layer of the intestine (Figures 4A, B). Under the connective tissue layer (Figure 4F), immunoreactivity in the acinar gland-like cells in the submuscular layer (Figures 4C, D, G) and the spindle-shaped epithelial cell (Figures 4E, H) was also detected.
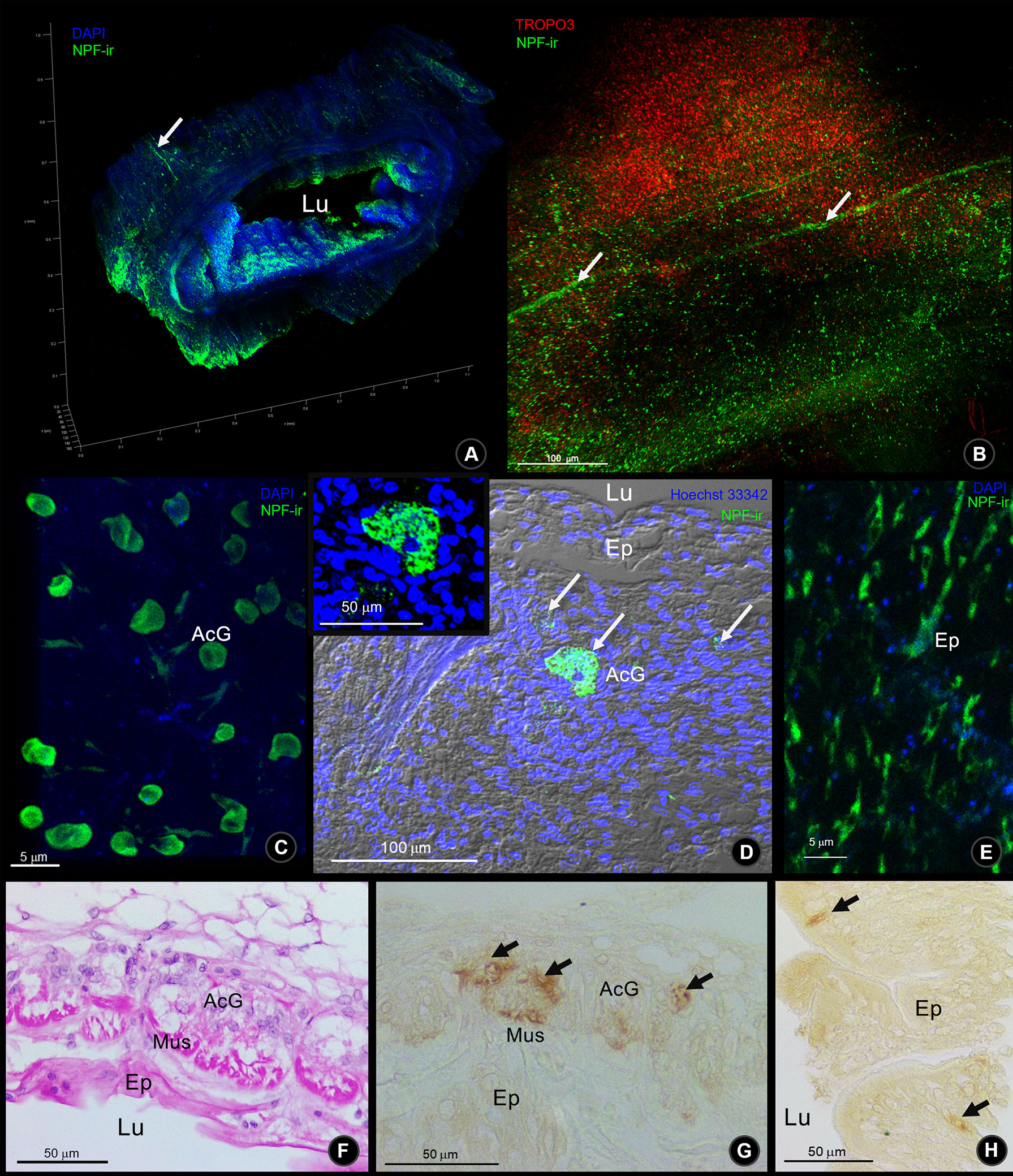
Figure 4 The Macro-NPF-ir detection in the intestine of S. olivacea. (A) Wholemount immunofluorescence of NPF-ir (green) counter-stained with DAPI revealed the overview of their localization. (B) The outer surface of intestinal serosa presented the NPF-ir in longitudinal nerve fibers. (C) Deep to the outer connective tissue, NPF-ir was localized in acinar glandular cells (arrows), revealed by wholemount immunofluorescence, and (D) by immunofluorescence in tissue section. (E) The NPF-ir localization presented in the spindle-shaped epithelial cells. (F) H&E staining of longitudinal section and (G) NPF-ir detection by immunohistochemistry (HRP detection) presented in acinar glandular cells of the subepithelial connective tissue layer (arrows). (H) Immunohistochemistry showed the positive signal of NPF localization in spindle-shaped epithelial cells (arrows).
Cloning and sequence analysis of S. olivacea NPF II cDNA
The cDNA sequence encoding S. olivacea NPF II was cloned using the PCR strategy, which is based on oligonucleotide primers corresponding with the NPF II conserved sequence of the other NPF from related crustacean species. A full length of S. olivacea NPF II was obtained, comprising 414 nucleotides in which 375 nucleotides were of open reading frames (ORF). The S. olivacea-NPF II encodes 124 deduced amino acids (Figure 5). This nucleotide sequence is available in the GenBank database (GenBank accession no. ON254657). A signal peptide for the S. olivacea-NPF II precursor was predicted, with cleavage occurring at positions A26 and A27. A 124-amino acid S. olivacea-NPF II prohormone was predicted to be cleaved into a putative NPF II mature peptide (KPDPTQLAQMADVIKYLHELDKYYSPMSRPSPRSAPGPASQIQALENTLKFLQLQELGKKYSHVTRPRFamide) (Figure 5).

Figure 5 Nucleotides corresponding with the deduced amino acid sequence of the full-length S. olivacea neuropeptide F II (Scyol-NPF II) are shown; within 414 nucleotides of Scyol-VIH cDNA sequence, and 375 were predicted to be an open reading frame. The open reading frame was translated into 124 deduced amino acids; small and capital letters indicate nucleotide and amino acid sequences, respectively. A black italic M letter represents a start codon, while an asterisk a stop codon. Signal peptide sequence of Scyol-NPF II is indicated by underlined italic letters. Black bold letter indicates the active form of NPF-II.
The full length of S. olivacea NPF II protein shares more than 89%–90% similarity to Scylla paramamosain NPFII and Callinectes arcuatus NPF II and matches 64%–75% with NPFI-II in other crustaceans (Figure 6, only partial alignment was shown). Moreover, S. olivacea NPF II shared 19%–22% to short-form NPF and 26%–30% to long-form NPF (Figure 6 and Supplement 5). Meanwhile, high conservation in NPF I-II amino acid composition mature form in crustaceans was also presented (Figure 7).
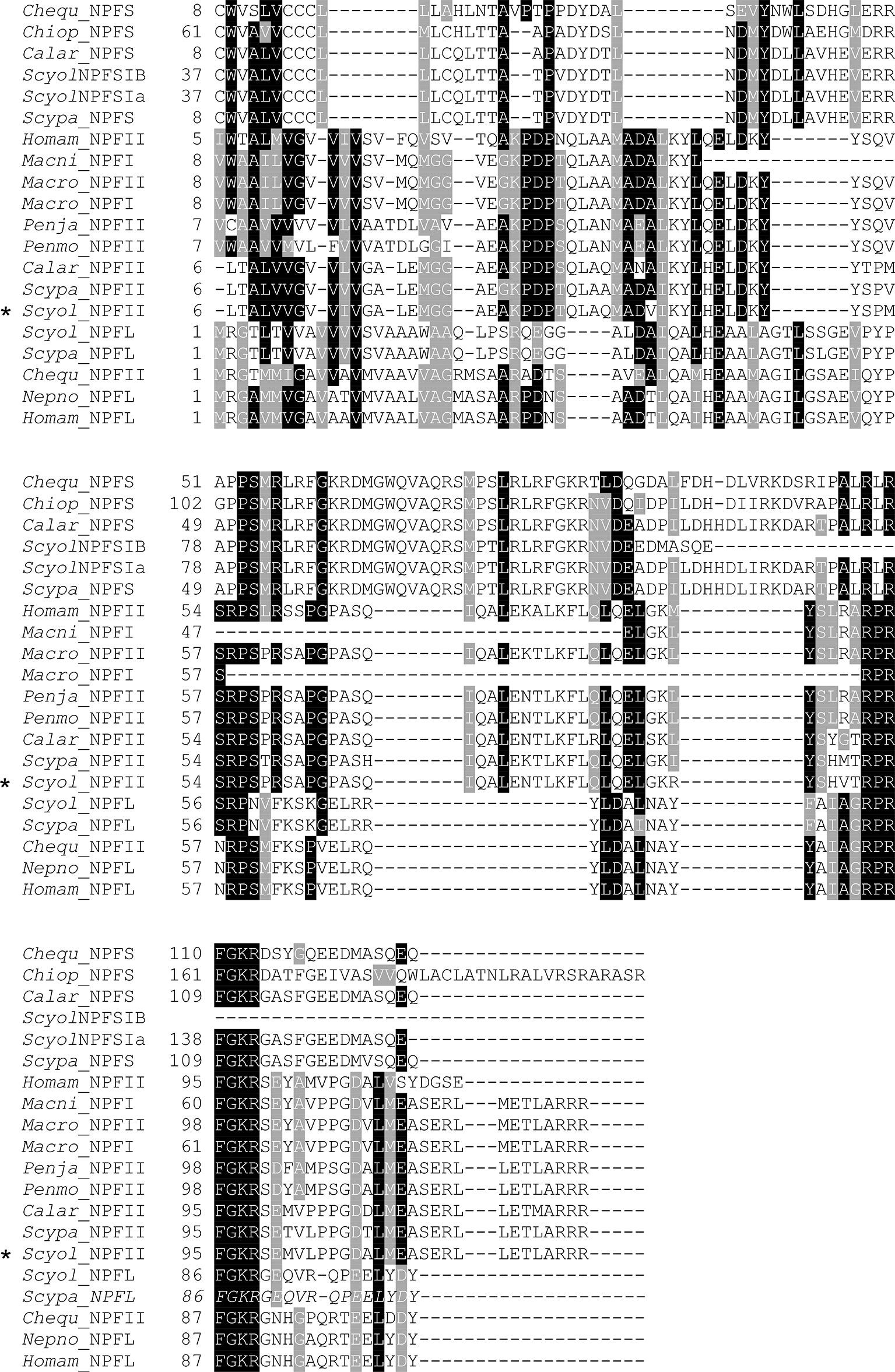
Figure 6 Comparative analysis of the S. olivacea NPF II protein sequence with other species. The multiple alignments of NPF II of S. olivacea proteins with NPF proteins in other species from crustaceans. Black shading in the multiple alignment indicates conserved amino acids while gray shading indicates similar amino acids. The asterisks indicate S. olivacea NPF II found in this study. Species abbreviation and GenBank accession numbers: Chequ, Cherax quadricarinatus (AWK57534.1, AWK57543.1); Chiop, Chionoecetes opilio (KAG0714693.1); Calar, Callinectes arcuatus (QPO25069.1, QPO25068.1); Homam, Homarus americanus (XP_042241562.1, XP_042234278.1); Macro, Macrobrachium rosenbergii (APP91265.1, APP91264.1); Macni, Macrobrachium nipponense (QJQ81519.1); Penja, Penaeus japonicus (XP_042870456.1); Penmo, Penaeus monodon (XM_037925377.1); Nepno, Nephrops norvegicus (QBX89063.1); Scypa, Scylla paramamosain (ALQ28587.1, ALQ28586.1, ALQ28574.1); Scyol, Scylla olivacea (ON254657, GDRN01070465, GDRN01012387, GDRN01012386).
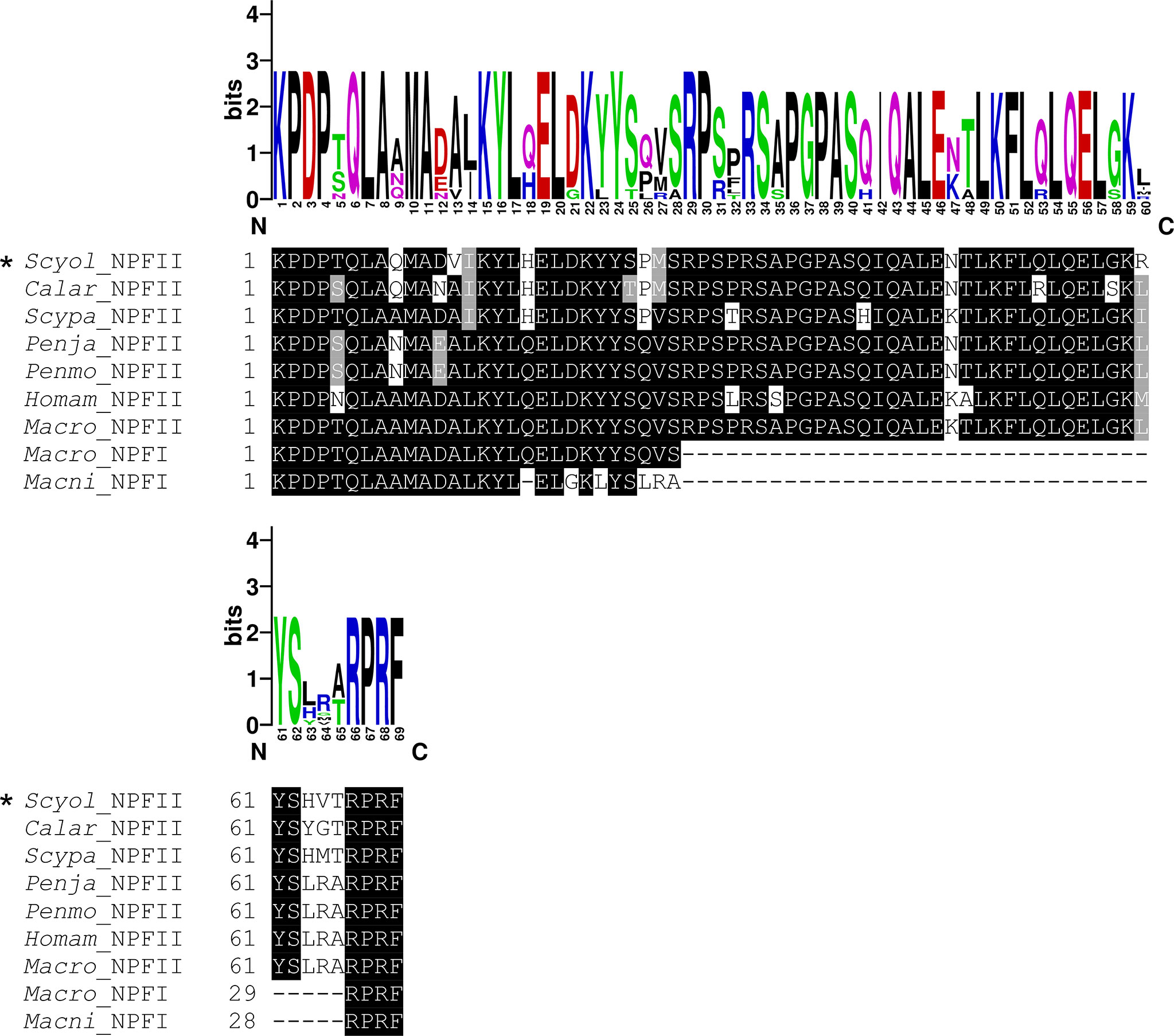
Figure 7 Comparative analysis of the active peptide of NPF II protein sequence with other species. The multiple alignments of NPF II of S. olivacea proteins with NPF proteins in other species from crustaceans. Black shading in the multiple alignments indicates conserved amino acids while gray shading indicates similar amino acids. The asterisks indicate S. olivacea NPF II found in this study. The sequence logo above the alignment shows the conservation of amino acid residues.
A phylogenetic analysis was constructed based on S. olivacea NPF II protein sequences and random and diverse examples within the NPF protein sequences. It indicated that S. olivacea NPF II was closely related to other homolog proteins in crustaceans. In addition, the phylogenetic tree displayed three distinct branches: (1) NPF I-II, (2) long-form NPF, and (3) short-form NPF (Figure 8).
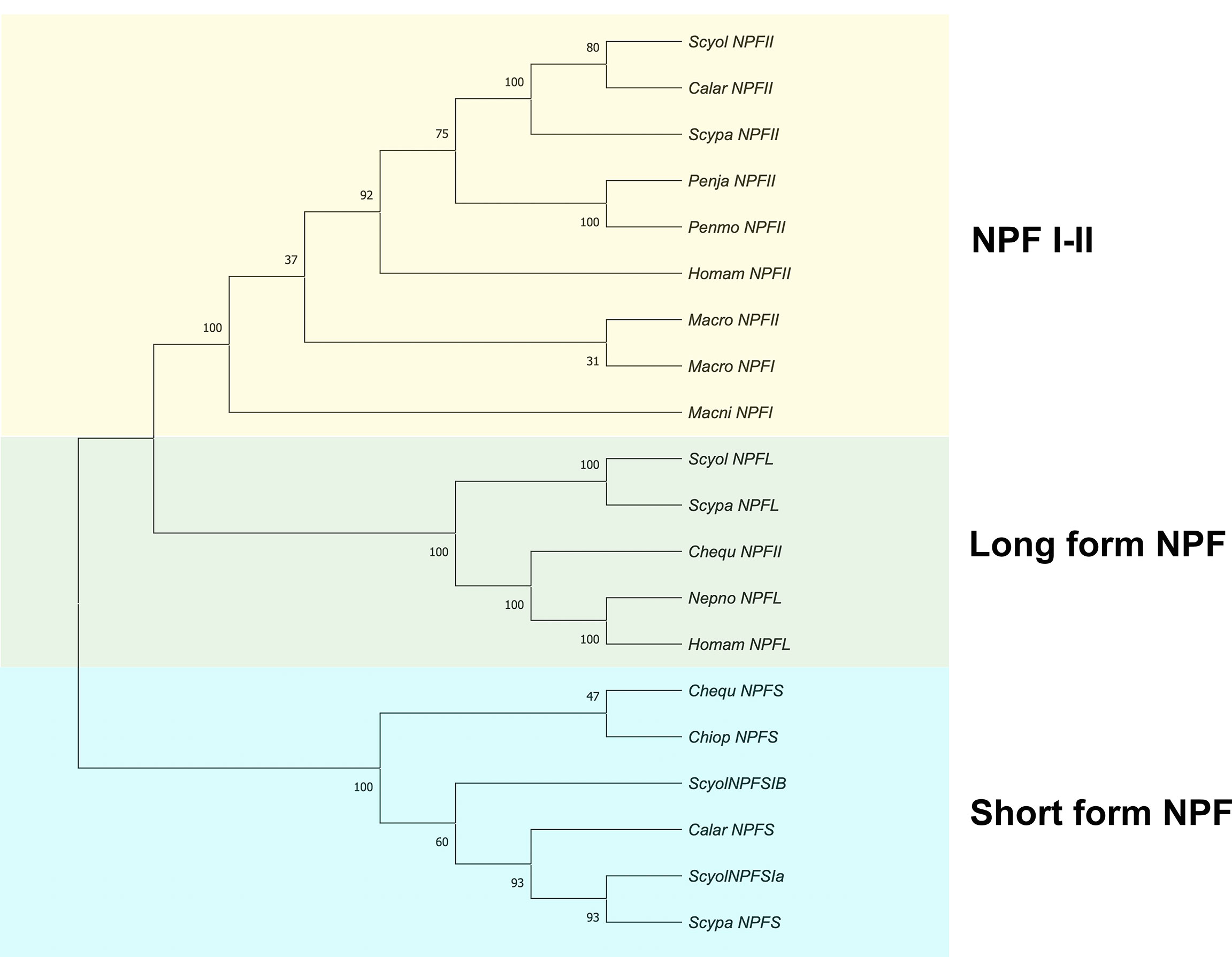
Figure 8 Phylogenetic tree analysis of NPF proteins from S. olivacea and other species from crustaceans. The phylogenetic tree was constructed based on the Neighbor-Joining method (number of amino acid substitution model). The number at the nodes represents bootstrap values at an approximate level of percent confidence from 1,000 bootstrap replicates. The asterisks indicate S. olivacea NPF II that was found in this study.
Tissue abundances of Scyol-NPF II mRNA transcript using RT-PCR
The relative abundance of S. olivacea NPF II mRNA transcript in different S. olivacea tissues was examined using RT-PCR and primers specific for S. olivacea NPF II. The S. olivacea NPF II transcripts were detected in the eyestalk, brain, ventral nerve cord, heart, intestine, and muscle of individual crabs at the expected size of 325 base pairs (Figure 9). There were no S. olivacea NPF II transcripts detected in other tissues examined (gill, stomach, and hepatopancreas) nor in the negative control samples.
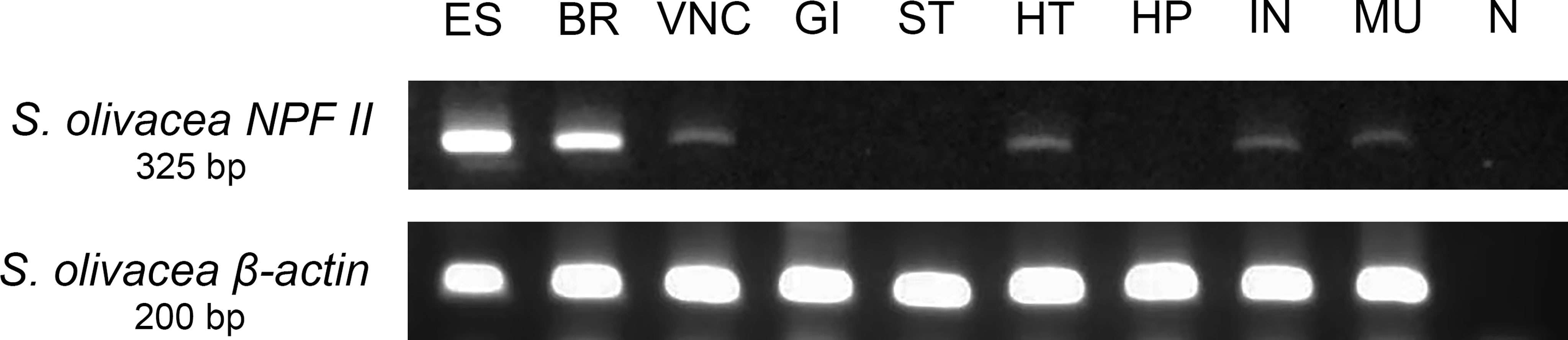
Figure 9 Tissue-specific relative abundances of S. olivacea NPF II mRNA transcript in mature female using RT-PCR; S. olivacea NPF II mRNA transcript is detected in the eyestalk (ES), brain (BR), ventral nerve cord (VNC), heart (HT), intestine (IN), muscle (MU), β-actin mRNA abundance was used as internal controls. ES, eyestalk; BR, brain; VNC, ventral nerve cord; OV, ovary; GI, gill; ST, stomach; HT, heart; HP, hepatopancreas; IN, intestine; MU, muscle and N negative control.
The specificity test of Scyol-NPF II peptide by dot-blot and immunohistochemistry analysis
The specificity binding between anti-Macro-NPF I antibody and the synthetic Scyol-NPF II peptide was meticulously conducted with two conventional methodologies, based on the antigen pre-absorption test. Firstly, the immunoblotting analysis of anti-Macro-NPF I antibody against the synthetic Scyol-NPF II peptide without pre-absorption presented NPF-ir. In contrast, the pre-absorption of the anti-Macro-NPF I antibody and the synthetic Scyol-NPF II peptide were able to diminish the NPF-ir (Figure 10A). As the negative control, BSA did not display any signal of either condition (Figure 10A). Secondly, the immunohistochemistry test showed that the anti-Macro-NPF I antibody had intense staining in the ON and the neuronal cluster 9/11 of S. olivacea tissue (Figure 10B). On the other hand, no NPF-ir was shown after the pre-absorption of the anti-Macro-NPF I antibody and the synthetic Scyol-NPF II peptide on the S. olivacea tissue were applied (Figure 10C).

Figure 10 Specificity test of the anti-MacroNPF antibody to the Scyol-NPF II synthetic peptide. (A) In dot-blot analysis, cross-reactivity tests of the anti-Macro-NPF I antibody showed immunopositivity to Scyol-NPF II synthetic peptide. The pre-absorption of the anti-Macro-NPF I antibody and the synthetic Scyol-NPF II peptide did not show immunopositivity. (B) Immunohistochemistry analysis, the positive signal of Macro-NPF-ir was detected the neurons of cluster 9/11 and olfactory neuropil, (C) no detection in the section treated with preabsorbed antibody.
Discussion
A hidden Scyol-NPF II had been initially revealed by unexpected observation via immunostaining with the Macro-NPF I antibody, according to the primary staining in the CNS of S. olivacea. Macro-NPF-ir was majorly detected in the CNS, including the neuron of clusters 2, 3, and 4 and neuropil ME, MI, MT, XO-SG complex of optic ganglion, a few neurons in cluster 6/7, AMPN, PMPN of the protocerebrum, neuronal cluster 9/11, neuropil ON and its accessory lobe, and OGTN of deutocerebrum, neuronal cluster 14/15, TN, AnN, and CN of tritocerebrum of the brain. This localization pattern was very similar to previous reports into M. rosenbergii, especially the extensive intensity of NPF-ir in the XO-SG complex and their fibers in the optic ganglion linking to the ON and associated neuronal cluster 9/11 of the brain via the PT (Thongrod et al., 2017; Tinikul et al., 2022). The distribution pattern showed similarity, especially in the optic ganglion; however, there is a small different pattern in the prawn’s brain. NPF-ir was found in a few neurons in cluster 6/7 of the mud crab, whereas its localization pattern was mainly in the small and medium-sized neurons of cluster 6 of the protocerebrum and mildly intense in the neurons of cluster 17 in M. rosenbergii (Thongrod et al., 2017; Tinikul et al., 2022). Moreover, our study revealed a distinct localization pattern of short NPF (sNPF) in S. paramamosain which was found in several neuronal clusters throughout the cerebral ganglion, including the neurons of cluster 10 examined by in situ hybridization (Xu et al., 2022). It is possible that the pattern of its localization might not be similar, according to the difference propose in the neuronal function or stages of release of each species (Thongrod et al., 2017; Tinikul et al., 2022; Xu et al., 2022). The most abundant NPF-ir throughout the CNS could suggest that this neuropeptide might be involved in the roles in olfactory modulation and endocrine signaling controls (Tinikul et al., 2022). Moreover, NPF-ir was localized in the neuronal clusters associated with SEG, TG, AG, and their neuropils of VNC of S. olivacea. Compared to M. rosenbergii NPF-ir, the localization patterns in TG and AG were quite similar to this study (Thongrod et al., 2019; Tinikul et al., 2022). Interestingly, NPF-ir was originally reported in CoG of the mud crab, S. olivacea, including the neuropils and associated neurons. The CoG connected between the brain and TG. It also contained motor neurons providing fibers to the stomatogastric ganglion which controlled ingesting and food processing of the foregut in lobster and crab (Kirby and Nusbaum, 2007; Marder and Bucher, 2007). According to the NPF-ir in CoG, it is possible that this neuropeptide might play a role associated with feeding regulation, besides the food sensation.
The antibody which had been used in this study was produced against the synthetic peptide NPF-I of the prawn M. rosenbergii as an immunogen for rabbit immunization. The epitope peptide sequence of the antibody was designed to bind with two long-forms of NPFs, including NPF-I and NPF-II isoforms of M. rosenbergii (Thongrod et al., 2017). The sequence of the Scyol-prepro-NPF reported by Christie (2016) and Macro-NPF I and II has been aligned based on multiple-sequence alignment and revealed that they were low identity in similarity. There is a high possibility that the Scyol-NPF which matches corresponds to the peptide sequence of Macro-NPF I remain uninvestigated. The Macro-NPF I-II peptide showed a similar identity to another species of mud crab, S. paramamosain NPF II, according to the report of Thongrod et al. (2017). Apart from the several forms of NPFs in this species (Christie, 2016), we also amplified the NPF-II transcript, which could be recognized by the anti-Macro-NPF I antibody and matched with the sequence of M. rosenbergii NPF I and NPF II. The sequence alignment results and phylogenic tree analysis of several crustacean species revealed that the putative S. olivacea NPF II nucleotide sequence from our study was a new form that had never been reported in this species before. It consisted of 414 nt with an open reading frame of 375 nt encoding a 124-deduce-translated peptide which showed close similarity to S. paramamosain NPF II and belonged to a clade of NPF II. Interestingly, it was quite distinct from previously reported isoforms of this species (Christie, 2016). This study showed three clades of the NPF family, including NPF I–II, long-form NPF, and short-form NPF. The phylogeny indicated that Scyol-NPF II belongs to the NPF I–II family sharing an identity with Macro-NPF I–II. When considered in the cluster of NPF II, this isoform is longer than NPF I with 37 amino acid sequences inserted in the middle region of peptide before the RPRFamide region (Christie et al., 2011; Thongrod et al., 2017). The Scyol-NPF II sequence showed a low similar identity (19%–22% similarity) from Scyol long-form (Scyol-NPFL) and Scyol short-form NPF (Scyol-NPFSIa and Scyol-NPFSIb), except RPRFamide and some active peptide regions (Christie, 2016). In S. paramamosain, two isoforms of NPFI-II and sNPF have been identified (Bao et al., 2015). They also suggested that sNPF may have a function involved in oocyte maturation in S. paramamosain (Bao et al., 2018), while NPF I in the crustaceans may be involved in feeding by increasing the growth rate in juvenile L. vannamei (Christie et al., 2011).
The NPF immunoreactivity was observed throughout the CNS by the wholemount immunofluorescence technique, which corresponded to the Scyol-NPF II expression by RT-PCR in this study. Scyol-NPF II was expressed in the ES, BR, VNC, and other non-neural tissues, similar to S. paramamosain NPF II, sNPF, and M. rosenbergii NPF I–II (Bao et al., 2015; Thongrod et al., 2017). Our study revealed the specific expression of NPF II in the intestine, without the detection in the stomach and hepatopancreas, while NPF II presented in intestine, stomach, and hepatopancreas in S. paramamosain (Bao et al., 2015). Interestingly, S. paramamosain NPF II expressed throughout the digestive organs in a similar way to M. rosenbergii, as reported by Thongrod et al. (2017). In contrast to our study, Scyol-NPF II was detected in the intestine of the alimentary tract without other related organs. It is possible that this isoform might perform a specific function in the CNS–intestinal axis. A few previous reports of D. melanogaster revealed that the localization of NPF was observed in the intestine, especially the proximal and middle parts of the midgut (Veenstra et al., 2008; Nässel, 2018; Hung et al., 2020; Yoshinari et al., 2021). The NPF was prominently expressed in the enteroendocrine cell (EEC) of the Drosophila midgut which co-expressed with tachykinin, neuropeptide-like precursor 2, and orcokinin (Hung et al., 2020). The NPF-expressed EEC of Drosophila midgut was reported to play a role as incretin-like EEC-derived hormones in mammals regulating the secretion of glucagon and insulins (Yoshinari et al., 2021). They also suggested that the function in the mediator regulatory for sugar-dependent metabolism regulated through glucagon-like and insulin-like hormones (Yoshinari et al., 2021). In this study, NPF-ir has been localized in the spindle-shaped epithelial cells of S. olivacea intestine. Although the population of gut epithelial cells of this species has not been clearly characterized, the NPF detected in the cells of this study may imply a similar function of the EEC to that of other species, such as acting as regulator for the metabolism of the intestine (Yoshinari et al., 2021). Moreover, NPF-ir also presented in the nerve fibers found at the outermost part of the intestine and interestingly in the acinar glandular-like cells in the submuscular layer of the intestinal wall in S. olivacea. Our results are the first to show the existence of NPF-ir in these structures. However, the structure and its proposed function remain unclear. The histology of the crayfish (P. clarkii) intestine showed the mucus acinar gland and bladder cells in subepithelial connective tissue, whose function were proposed for packaging, lubrication of fecal strand, and shock absorbers to prevent damage of constituent neurons (To et al., 2004). However, the pattern of neuropeptide from the brain–gut axis was dominantly revealed in vertebrated species, including Nile tilapia, Oreochromis niloticus; the NPY immunoreactivity was found in enteroendocrine epithelial cells and myenteric plexus of the intestine (Pereira et al., 2017). This pattern is similar to the localization of S. olivacea. It is possible that the acinar gland-like cells found in the mud crab’s intestinal wall might act as myenteric plexus, which was not identified in terms of structure and function in crustacean species. This pattern supposed that NPF might be involved in the function similar to the human brain–gut and gut–brain axes, with bidirectional communication between the gut and brain (Holzer et al., 2012).
Taken together, the localization of NPF-ir in the mud crab (S. olivacea) throughout the CNS tissues, including eyestalk, brain, and ventral nerve cord, was revealed to correspond to probing with the Macro-NPF I antibody. Apart from the CNS, NPF-ir in the nerve fibers, the epithelial cells, and acinar glandular-like cells may perform as the enteroendocrine cell in the intestinal tract of this species. This study is the first report to show a hidden form of NPF-II to be investigated in S. olivacea by molecular cloning. Our results correspond to the existence, distribution, and expression similar to other crustaceans. Few differences between our study and previous reports were obvious in the localization in the intestinal tissues. According to the expression of NPF, this study may imply the function of NPF which could play a role as the mediator for gut modulation and might be involved in metabolic control in the CNS–gut axis for this species, as well as function in other invertebrate and vertebrate species. This research may provide knowledge about the neuroendocrine axis which may be essential for mud crab aquaculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
TK and NK designed the experiment, analyzed the data, and wrote the manuscript. TK, NK, SD, and SS performed the experiments. TK, NK, RV, CC, and PS conceptually designed the experiment, provided the experimental tools, and made the manuscript revisions. All authors contributed to the article and approved the submitted version.
Funding
This study is officially supported by the Mahidol University (Fundamental Fund: Basic Research Fund, the fiscal year 2021) to TK (Grant no. BRF1-A26/2564) and the supplementary and facility support grants from the Faculty of Science, Mahidol University. This present work was also supported by the Thammasat University Research Unit in Biotechnology and its application of aquatic animals.
Acknowledgments
The authors are especially grateful to Mahidol University, Fundamental Fund: Basic Research Fund, the fiscal year 2021, to TK, the supplementary and facility support grants from the Faculty of Science, Mahidol University and Thammasat University Research Unit in Biotechnology and its application of aquatic animals. The authors would like to thank Dr. Piyachat Chansela and Dr. Sirorat Thongrod for kindly providing the NPF antibody in the study and Mrs. Kornchanook Jaiboon for her kind assistance. The authors would also like to express our gratitude to Asst. Prof. Dr. Jirawat Saetan for his antibody specificity test suggestion. The authors thank the Center of Nanoimaging (CNI), Faculty of Science, Mahidol University, for instrumental and microscopic support. Finally, we would like to thank Mr. Wattapong Pinyo for his kindness in assisting in microscopic use. Finally, the authors are grateful to Mr. John Tucker, MA in Language Testing, University of Lancaster, for his kind help in the English correction.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.951648/full#supplementary-material
References
Bao C., Yang Y., Huang H., Ye H. (2015). Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: transcriptomic analysis and expression profiles during vitellogenesis. Sci. Rep. 5, 17055. doi: 10.1038/srep17055
Bao C., Yang Y., Huang H., Ye H. (2018). Inhibitory role of the mud crab short neuropeptide f in vitellogenesis and oocyte maturation via autocrine/paracrine signaling. Front. Endocrinol. 9. doi: 10.3389/fendo.2018.00390
Christie A. E. (2016). Prediction of Scylla olivacea (Crustacea; brachyura) peptide hormones using publicly accessible transcriptome shotgun assembly (TSA) sequences. Gen. Comp. Endocrinol. 230-231, 1–16. doi: 10.1016/j.ygcen.2016.03.008
Christie A. E., Cashman C. R., Brennan H. R., Ma M., Sousa G. L., Li L., et al. (2008). Identification of putative crustacean neuropeptides using in silico analyses of publicly accessible expressed sequence tags. Gen. Comp. Endocrinol. 156, 246–264. doi: 10.1016/j.ygcen.2008.01.018
Christie A. E., Chapline M. C., Jackson J. M., Dowda J. K., Hartline N., Malecha S. R., et al. (2011). Identification, tissue distribution and orexigenic activity of neuropeptide f (NPF) in penaeid shrimp. J. Exp. Bio 214, 1386–1396. doi: 10.1242/jeb.053173
Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Cui W., Guan M., Sadek A., Fangchun W., Qingyang W., Huaqiang T., et al. (2021). Construction of a genetic linkage map and QTL mapping for sex indicate the putative genetic pattern of the F1 hybrid Scylla (Scylla serrata ♀ × s. paramamosain ♂). Aquaculture 545, 737222. doi: 10.1016/j.aquaculture.2021.737222
Cui H., Zhao Z. (2020). Structure and function of neuropeptide f in insects. J. Integ. Agri. 19, 1429–1438. doi: 10.1016/S2095-3119(19)62804-2
Duangprom S., Ampansri W., Suwansa-ard S., Chotwiwatthanakun C., Sobhon P., Kornthong N. (2018). Identification and expression of prostaglandin e synthase (PGES) gene in the central nervous system and ovary during ovarian maturation of the female mud crab, Scylla olivacea. Anim. Reprod. Sci. 198, 220–232. doi: 10.1016/j.anireprosci.2018.09.022
Duangprom S., Kornthong N., Suwansa-ard S., Srikawnawan W., Chotwiwatthanakun C., Sobhon P. (2017). Distribution of crustacean hyperglycemic hormones (CHH) in the mud crab (Scylla olivacea) and their differential expression following serotonin stimulation. Aquaculture 468, 481–488. doi: 10.1016/j.aquaculture.2016.11.008
Fadda M., Hasakiogullari I., Temmerman L., Beets I., Zels S., Schoofs L. (2019). Regulation of feeding and metabolism by neuropeptide f and short neuropeptide f in invertebrates. Front. Endocrinol. 10. doi: 10.3389/fendo.2019.00064
Farhadi A., Lv L., Song J., Zhang Y., Ye S., Zhang N., et al. (2022). Whole-transcriptome RNA sequencing revealed the roles of chitin-related genes in the eyestalk abnormality of a novel mud crab hybrid (Scylla serrata ♀ × s. paramamosain ♂). Int. J. Biol. Macromol. 208, 611–626. doi: 10.1016/j.ijbiomac.2022.03.135
Holzer P., Reichmann F., Farzi A. (2012). Neuropeptide y, peptide YY and pancreatic polypeptide in the gut-brain axis. Neuropeptides 46, 261–274. doi: 10.1016/j.npep.2012.08.005
Hung R. J., Hub Y., Kirchner R., Liu Y., Xu C., Comjean A., et al. (2020). A cell atlas of the adult drosophila midgut. Proc. Natl. Acad. Sci. U.S.A. 117, 1514–1523. doi: 10.1073/pnas.1916820117
Ikhwanuddin M., Azmie G., Juariah H. M., Zakaria M. Z., Ambak M. A. (2011). Biological information and population features of mud crab, genus Scylla from mangrove areas of Sarawak, Malaysia. Fish. Res. 108, 299–306. doi: 10.1016/j.fishres.2011.01.001
Islam M. S., Kodama K., Kurokora H. (2010). Ovarian development of the mud crab Scylla paramamosain in a tropical mangrove swamps, Thailand. J. Sci. Res. 2, 380–389. doi: 10.3329/jsr.v2i2.3543
Kirby M. S., Nusbaum M. P. (2007). Central nervous system projections to and from the commissural ganglion of the crab cancer borealis. Cell Tissue Res. 328, 625–637. doi: 10.1007/s00441-007-0398-2
Kornthong N., Duangprom S., Suwansa-ard S., Saetan J., Phanaksri T., Songkoomkrong S., et al. (2019). Molecular characterization of a vitellogenesis-inhibiting hormone (VIH) in the mud crab (Scylla olivacea) and temporal changes in abundances of VIH mRNA transcripts during ovarian maturation and following neurotransmitter administration. Anim. Reprod. Sci. 208, 106122. doi: 10.1016/j.anireprosci.2019.106122
Kruangkum T., Chotwiwatthankun C., Vanichviriyakit R., Tinikul Y., Anuracpreeda P., Wanichanon C., et al. (2013). Structure of the olfactory receptor organs, their GABAergic neural pathways, and modulation of mating behavior, in the giant freshwater prawn, Macrobrachium rosenbergii. Microsc. Rese Tech. 76, 572–587. doi: 10.1002/jemt.22202
Marder E., Bucher D. (2007). Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu. Rev. Physiol. 69, 291–316. doi: 10.1146/annurev.physiol.69.031905.161516
Nässel D. R. (2018). Substrates for neuronal cotransmission with neuropeptides and small molecule neurotransmitters in drosophila. Front. Cell Neurosci. 12. doi: 10.3389/fncel.2018.00083
Nässel D. R., Wegener C. (2011). A comparative review of short and long neuropeptide f signaling in invertebrates: any similarities to vertebrate neuropeptide y signaling? Peptides 32, 1335–1355. doi: 10.1016/j.peptides.2011.03.013
Nooseng S. (2015). “Status of mud crab industry in Thailand,” in Proceedings of the international seminar-workshop on mud crab aquaculture and fisheries management, 10-12 April 2013, Tamil nadu, India. Eds. Quinitio E. T., Parado-Estepa F. D., Raj Y.C.T. S., Mandal A. (Tamil Nadu, India: Rajiv Gandhi Centre for Aquaculture (MPEDA), 37–43.
Onken H., Moffett S. B., Moffett D. F. (2004). The anterior stomach of larval mosquitoes (Aedes aegypti): effects of neuropeptides on transepithelial ion transport and muscular motility. J. Exp. Biol. 207, 3731–3739. doi: 10.1242/jeb.01208
Pereira R. T., de Freitas T. R., de Oliveira I. R. C., Costa L. S., Vigliano F. A., Rosa P. V. (2017). Endocrine cells producing peptide hormones in the intestine of Nile tilapia: distribution and effects of feeding and fasting on the cell density. Fish. Physiol. Biochem. 43, 1399–1412. doi: 10.1007/s10695-017-0380-1
Shelley C. (2008). “Capture-based aquaculture of mud crabs (Scylla spp.),” in Capture-based aquaculture. global overview, FAO fisheries technical paper. no. 508. Eds. Lovatelli A., Holthus P. F. (Rome: FAO), 255–269.
Shen P., Cai H. N. (2001). Drosophila neuropeptide f mediates integration of chemosensory stimulation and conditioning of the nervous system by food. J. Neurobiol. 47, 16–25. doi: 10.1002/neu.1012
Stecher G., Tamura K., Kumar S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239. doi: 10.1093/molbev/msz312
Suwansa-Ard S., Thongbuakaew T., Wang T., Zhao M., Elizur A., Hanna P. J., et al. (2015). In silico neuropeptidome of female Macrobrachium rosenbergii based on transcriptome and peptide mining of eyestalk, central nervous system, and ovary. PloS One 10, e123848. doi: 10.1371/journal.pone.0123848
Thongrod S., Changklungmoa N., Chansela P., Siangcham T., Kruangkum T., Suwansa-Ard S., et al. (2017). Characterization and tissue distribution of neuropeptide f in the eyestalk and brain of the male giant freshwater prawn, Macrobrachium rosenbergii. Cell Tissue Res. 367, 181–195. doi: 10.1007/s00441-016-2538-z
Thongrod S., Wanichanon C., Sobhon P. (2019). Distribution of neuropeptide f in the ventral nerve cord and its possible role on testicular development and germ cell proliferation in the giant freshwater prawn, macrobrachium rosenbergii. Cell Tissue Res. 376, 471–484. doi: 10.1007/s00441-019-02999-8
Tinikul Y., Kruangkum T., Tinikul R., Sobhon P. (2022). Comparative neuroanatomical distribution and expression levels of neuropeptide f in the central nervous system of the female freshwater prawn, Macrobrachium rosenbergii, during the ovarian cycle. J. Comp. Neurol. 530, 729–755. doi: 10.1002/cne.25241
To T. H., Brenner T. L., Cavey M. J., Wilkens J. L. (2004). Histological organization of the intestine in the crayfish procambarus clarkii. Acta Zool. (Stockholm) 85, 119–130. doi: 10.1111/j.0001-7272.2004.00164.x
Veenstra J. A., Agricola H. J., Sellami A. (2008). Regulatory peptides in fruit fly midgut. Cell Tissue Res. 334, 499–516. doi: 10.1007/s00441-008-0708-3
Wu Y., He H., Cheng Z., Bai Y., Ma X. (2019). The role of neuropeptide y and peptide YY in the development of obesity via gut-brain axis. Curr. Protein Pept. Sci. 20, 750–758. doi: 10.2174/1389203720666190125105401
Wu Q., Wen T., Lee G., Park J. H., Cai H. N., Shen P. (2003). Developmental control of foraging and social behavior by the drosophila neuropeptide y-like system. Neuron 39, 147–161. doi: 10.1016/s0896-6273(03)00396-9
Xu Z., Wei Y., Huang H., Guo S., Ye H. (2022). Immunomodulatory role of short neuropeptide f in the mud crab Scylla paramamosain. Dev. Comp. Immunol. 126, 104260. doi: 10.1016/j.dci.2021.104260
Yoshinari Y., Kosakamoto H., Kamiyama T., Hoshino R., Matsuoka R., Kondo S., et al. (2021). The sugar-responsive enteroendocrine neuropeptide f regulates lipid metabolism through glucagon-like and insulin-like hormones in Drosophila melanogaster. Nat. Commun. 12, 4818. doi: 10.1038/s41467-021-25146-w
Keywords: mud crab, neuropeptide F, brain-gut axis, enteroendocrine cell, feeding control
Citation: Kruangkum T, Duangprom S, Songkoomkrong S, Chotwiwatthanakun C, Vanichviriyakit R, Sobhon P and Kornthong N (2022) Discovery of a hidden form of neuropeptide F and its presence throughout the CNS–gut axis in the mud crab, Scylla olivacea. Front. Mar. Sci. 9:951648. doi: 10.3389/fmars.2022.951648
Received: 24 May 2022; Accepted: 22 July 2022;
Published: 15 August 2022.
Edited by:
Hongyu Ma, Shantou University, ChinaCopyright © 2022 Kruangkum, Duangprom, Songkoomkrong, Chotwiwatthanakun, Vanichviriyakit, Sobhon and Kornthong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Napamanee Kornthong, napamaneenatt@gmail.com
 Thanapong Kruangkum
Thanapong Kruangkum Supawadee Duangprom
Supawadee Duangprom Sineenart Songkoomkrong3
Sineenart Songkoomkrong3  Charoonroj Chotwiwatthanakun
Charoonroj Chotwiwatthanakun Rapeepun Vanichviriyakit
Rapeepun Vanichviriyakit Napamanee Kornthong
Napamanee Kornthong