Probiotic properties of Bacillus strains isolated from the gastrointestinal tract against pathogenic Vibriosis
- 1Laboratory of Microbial Biotechnology and Engineering Enzymes (LBMIE), Center of Biotechnology of Sfax (CBS), University of Sfax, Sfax, Tunisia
- 2Astrum Biotech, Business Incubator, Center of Biotechnology of Sfax (CBS), University of Sfax, Sfax, Tunisia
Vibriosis is one of the major diseases leading to massive fish mortality. Probiotics may provide a potential alternative method to protect fish from pathogens and to promote a balanced environment minimizing the use of antibiotics and chemotherapy. The aims of this study were to (i) isolate and purify marine spore-former strains from Sardine and shrimp intestine, (ii) screen for bacteria with potential probiotic properties, and (iii) carry out their in vitro safety assessment using a subtractive procedure. Among 108 spore-former strains, five strains exhibited a strong antibacterial activity against Vibriosis such as Vibrio harveyi and Vibrio anguillarum. These selected strains were unaffected by high-temperature and gastrointestinal conditions; produced amylase, protease, and lipase activities; and showed high percentages of auto-aggregation and co-aggregation with pathogens, as well as a strong adhesion to fish mucus. Partial 16S rDNA gene sequencing and MALDI-TOF MS revealed that isolates are Bacillus amyloliquefaciens or Bacillus subtilis. All of them were susceptible to antibiotics, while hydrolic enzymes and virulence factors were not detected for B. subtilis S17. In conclusion, based on their proprieties and their safety assessment, B. subtilis S17 could serve as a potential probiotic candidate for aquaculture.
1 Introduction
Intensive aquaculture is known to cause stressful conditions related to stocking densities and high feeding. Such practices enhance the probability of disease outbreaks mainly at larval and early fry stages. Outbreaks are habitually caused by a wide range of pathogenic bacteria. Vibriosis is one of the major diseases leading to poor growth, low immunity, and high mortality and therefore massive economic losses for the aquaculture sector (Meidong et al., 2017; Kaktcham et al., 2018; Ghanei-Motlagh et al., 2020; Makridis et al., 2021). Several Vibrio species have been concerned with marine animals’ health problems. Recent studies showed that Vibrio alginolyticus, V. harveyi, and V. parahaemolyticus are the most potent species infecting farms (Daniels and Shafaie, 2000; Yılmaz et al., 2021). To sort out these problems, traditional methods such as vaccines and antibiotics are widely used. Nevertheless, the widespread use of antibiotics leads to the appearance and emergence of resistant bacteria and affect the aquatic environment (water and sediment contamination) (Caruffo et al., 2015; Medina et al., 2020; Makridis et al., 2021). Recently, probiotics have emerged as a good and effective alternative to ban or minimize the antibiotic use and control diseases (Balcazar et al., 2007; Meidong et al., 2017; Emam and Dunlap, 2020). Probiotics are defined as live microorganisms which beneficially affect the health of the host when consumed in adequate concentrations (Fuller, 1989; FAO/WHO/OIE, 2006; Pinto et al., 2020). Their potential benefits on the host are attributed to improve the immune system and enhance digestion and growth promotion by producing enzymes. Other beneficial effects include water remediation and secretion of inhibitory substances which confer resistance to intestinal pathogens and prevent their intestinal adhesion, and lead to better health properties (Verschuere et al., 2000; Balcázar et al., 2006; Balcazar et al., 2007; Fernandes and Kerkar, 2019; Medina et al., 2020; Khan et al., 2021). A large number of bacteria are classified as probiotics, but the most widely used in aquaculture are Lactobacillus and Bacillus. However, Bacillus species have attracted much more attention due to non-toxic and non-pathogenic compound production and their sporulation ability, which gives them protection against stressed conditions (heat, gastrointestinal tract (GIT) compared to other probiotics (Barbosa et al., 2005; Cao et al., 2019; Kuebutornye et al., 2019; Emam and Dunlap, 2020; Amoah et al., 2021). To exert and endure the benefits of probiotics within the host, several properties should be investigated for probiotic strain screening, including antagonism against fish pathogens, being kept alive in GIT, resistance to storage conditions, secretion of exogenous enzymes, and being safe (Fuller, 1989; Muñoz-Atienza et al., 2014; Cao et al., 2019; Zhou et al., 2019; Ghanei-Motlagh et al., 2020). Generally, probiotics are isolated from the indigenous and exogenous microbiota of aquatic animals (Balcázar et al., 2006; do Vale Pereira et al., 2017). However, the search for a putative probiotic should continue as the increase demand for sustainable and environment-friendly aquaculture.
In this study, we used Sardine and shrimp intestine, an untapped source of novel microorganism, as a source of potential probiotic bacteria. Hence, we have isolated, selected, and identified potential probiotic bacteria for aquaculture based on subtractive screening by means of several physiologic criteria and safety assessment.
2 Materials and methods
2.1 Pathogen collection and culture conditions
All Vibrio used as indicators (Vibrio anguillarum CECT4344; V. alginolyticus; V. fischeri; V. harveyi Lg 10/6, Lg 14/01, Lg 26/01, Lg 48/01, Lg 13/04, Lg 34/04, and Lg 35/03; V. parahaemolyticus; V. proteolyticus; V. vulnificus, and V. splendidus provided by The Microbiology Department of the University of Malaga) were isolated from diseased fish. They were potent causative agents of fish infection. Each strain was cultured in Tryptone Soya Agar (TSA) (Oxoid Ltd., Hampshire, UK) supplemented with NaCl 1.5% (w/v) at 28°C. Stock cultures in Tryptone Soy Broth (TSB) were stored at -80°C and -20°C in 1.5% NaCl with 15% glycerol to provide stable inoculum throughout the study (Midhun et al., 2018).
2.2 Sampling procedure and isolation of spore-forming bacteria
Bacteria used in this study were isolated from intestines of Sardine (Sardina pilchardus) and shrimp (Penaeus kerathurus). Intestines were washed with sterile saline solution, and content was aseptically removed. One gram from each intestine sample was homogenized in 9 ml phosphate-buffered saline (PBS, pH = 7.2). Selection of spore-forming isolates was by heat treatment. The homogenates were diluted in buffered peptone water (up to 10−3) and incubated at 65°C for 30 min. One hundred-microliter aliquots of appropriate serial dilutions were spread onto Difco sporulation medium (DSM) for 48h at 28°C. Colonies obtained were picked and purified. Pure isolates in TSB were stored in 1.5% NaCl with 15% glycerol at -20°C and -80°C until further routine use (Barbosa et al., 2005).
All isolates were then investigated for catalase activity by the suspension of a fresh single colony in 3% of hydrogen peroxide. Air bubble production was considered as a positive result.
2.3 Antimicrobial activity
2.3.1 Colony overlay method
The preliminary screening for antimicrobial activity of spore-forming isolates was done by the colony overlay method described by Alonso et al. (2019). Both indicators and isolates were grown in TSB containing 1.5% NaCl and incubated overnight at 28°C. In order to form a bacterial lawn, 100 μl Vibrio spp. Culture (105 CFU/ml) was spread in TSA plates using a sterile cotton-tipped swab. Drops of isolate cultures were dispensed onto the plates once dried. After incubation at 28°C for 24 h, antimicrobial activity was assessed by observing the presence of inhibition zones around colonies and represented as scores as described by Midhun et al. (2017). Based on cumulative scores, promising antagonistic spore-forming bacteria against indicators were selected. Further, the strong Vibrio inhibitory spore-forming bacteria were confirmed through cell-free supernatants (CFSs).
2.3.2 Extracellular antimicrobial assay
The extracellular antimicrobial activity of cell-free supernatants was determined using a microtiter plate and an agar well diffusion test (ADT). Briefly, selected strains were cultured in TSB containing 1.5% NaCl incubated at 28°C for 24 h at 150 rpm. CFS was obtained by centrifugation of overnight cultures (107 CFU/ml) at 8,000×g at 4°C for 20 min and filter sterilized through 0.22-μm pore-size filters.
The inhibitory activity was evaluated in a 96-well microtiter plate by inoculation of pathogenic bacteria culture (10 μl, 105 CFU/ml) with TSB fresh medium (150 μl) and sterilized CFS (50 μl) in each well. Controls were inoculated with pathogenic bacteria only in 200 μl fresh TSB. The absorbance at 600 nm was used to measure the inhibition efficiency (IE) during 24 h by the following formula:
where IE<1 indicates inhibition of pathogenic bacteria by isolates, IE = 1 indicates no inhibition, and IE >1 indicates growth promotion (Mukherjee and Ghosh, 2016).
Besides, antibacterial activity using ADT was assayed as described by Cintas et al. (1995) with some modifications. Plates were seeded with indicators (100 μl, 105 CFU/ml). Fifty microliters of CFS was placed into wells (6 mm of diameter) cut in TSB with 1.5% NaCl and 0.8% agar (w/v). After 2 h at 4°C, the plates were incubated under 24 h at 28°C then antimicrobial activity was defined by the presence of a clear inhibitory zone around the wells. Further, neutralization was obtained using 1 N sodium hydroxide to pH = 7. To determine the nature and thermostability of the antimicrobial compounds, the supernatant was treated with proteinase K at 10 mg/ml (AppliChem GmbH, Darmstadt, Germany) and then heated at 100°C for 10 min. After treatments, samples were assayed for residual antimicrobial activity by an ADT using Vibrio anguillarum CECT4344 and V. harveyi Lg26/01 as indicators.
2.4 Preparation of vegetative cells, spores, and sporulation efficiency
For the preparation of vegetative cells, each selected strain was grown in TSB with 1.5% NaCl and taken from logarithmic-phase cells.
Sporulation of isolates was induced by exhaustion of nutrients in DSM. In fact, overnight cultures of selected strains in TSB with 1.5% NaCl were inoculated to DSM broth and incubated 28°C at 150 rpm for 24 and 48 h. After inoculation, cultures were diluted and plated in TSA in order to determine viable and heat-resistant cells before and after heat treatment (80°C, 20 min), respectively. Sporulation efficiency was determined by quantifying viable cells before and after heating (Barbosa et al., 2005; Prieto et al., 2014; Zhou et al., 2019; Santos et al., 2021).
2.5 Screening of probiotic properties of spore-forming isolates
2.5.1. Evaluation of extracellular enzymes
The extracellular enzyme production of selected bacteria was performed by plate assay according to Hmani et al. (2017) with some modifications. Protease and amylase production was determined using Luria-Bertani (LB) agar plates supplemented with 1% skimmed milk and 1% starch, respectively. Culture plates were spot inoculated and incubated for 24 h at 28°C. The clear zone around the colonies revealed a proteinase production. After plates were flooded with iodine, yellow discoloration appeared, indicating amylase production. For lipase activity, strains were spotted onto LB plates containing 1% olive oil and 1% rhodamine. Lipase activity was observed by zone of clearance surrounding colonies.
2.5.2 Biofilm production
The ability to produce biofilm was determined by the crystal violet assay as described by Midhun et al. (2017) with modifications. Selected strains were inoculated into 10 ml of LB broth containing 1% glucose and incubated at 28°C for 24 h. Cultures were diluted to an OD600 of ~0.1. Each well of a sterile 96-well flat-bottom polystyrene plate (orange scientific) was filled with 200 μl of diluted culture. Escherichia coli DH5α and Staphylococcus aureus ATCC 6538 were used as negative and positive biofilm-forming strains, respectively, and sterile LB broth was considered as blank to check the sterility and non-specific attachment. After incubation, excess medium from each well was removed by tapping. The wells were washed twice with 250 μl of PBS (pH = 7.2) to eliminate floating bacteria. The adherent bacteria were fixed at 60°C for 1 h and stained with 150 μl of 0.2% crystal violet prepared in 20% EtOH (v/v). The stained bound bacteria were released by adding 200 μl of glacial acetic acid. After incubation for 1 h at room temperature, the absorbance at 570 nm was determined using a microplate reader. The wells of isolates in which OD values are higher than those in blank wells were considered to be biofilm producers.).
2.5.3 Resistance to high temperature, pH, and bile salts
The resistance of selected strains to high temperatures required by the fish feed process was assessed according to Kuebutornye et al. (2020) with modifications. Briefly, an overnight culture was harvested. The pellets were resuspended in PBS and exposed to elevated temperatures (80°C, 90°C, and 100°C) for 5 and 10 min, respectively. Viable counts were determined by the spread plate method after incubation at 28°C for 24 h.
The tolerance of vegetative cells and spores to low pH and bile salt was assayed as described by Barbosa et al. (2005) and Hmani et al. (2017) with modifications. Essentially, ~108 to 109 bacterial cells ml-1 were resuspended in TSB broth containing bile salts (0%–5%, sodium cholate 50%, sodium deoxycholate 50%) (Bio Basic, Markham, Canada) or in TSB broth adjusted to pH 1, 2, 3, and 7.3 (control) with concentrated HCl. Aliquots were taken immediately and after 3 h for low pH and after 6 h for bile salt tolerance. Viable counts were determined by the spread plate method on TSA after incubation at 28°C for 24 h.
2.5.4 Auto-aggregation and co-aggregation assays
Auto-aggregation assay was performed according to Meidong et al. (2018). Overnight cultures grown were centrifuged, washed twice, and resuspended in PBS followed by turbidity measurement to obtain OD600~1(A0). The bacterial suspensions were vortexed for 10 s and kept undisturbed at 28°C for 5 h and after 24 h. Then each hour OD600 of the upper suspensions (At) was measured. The auto-aggregation percentage (%Agg) was expressed as:
where At represents the OD600 at time t = 1, 2, 3, 4, 5, and 24 h and A0 the OD600 at t = 0.
In the co-aggregation assay, the suspension of isolated strains and fish pathogens was prepared similarly to the auto-aggregation analysis above. Equal volumes of tested strains and pathogens were mixed and incubated at 28°C for 6 h. The OD600 of the mixtures and controls (unmixed cultures of tested strains and pathogens) were measured after incubation. Co-aggregation (Co-agg) was calculated as:
where Apat and Aisolate represent the OD600 of each bacterial suspension alone and Amix represents the OD600 of mixed suspension pathogen and isolated strains (Meidong et al., 2018).
2.5.5 Adhesion assay in vitro
2.5.5.1 Mucus preparation and characterization
Mucus was isolated from the skin and intestine of gilt-head bream (Sparus aurata) according to Balcázar et al. (2008). The skin mucus was collected by scraping the dorsolateral surfaces of fish using a cell scarper. The intestine was removed and scraped to collect intestinal mucus. All mucus were homogenized in PBS and centrifuged three times at 120,000× g for 15 min at 4°C to remove cellular materials and particulate. Then the samples were adjusted to a protein concentration of 1 mg/ml in PBS. The protein concentration was determined according to Beltrán et al. (2017) and Midhun et al. (2018). The mucus preparations were filter sterilized by a 0.22-µm-size Millipore filter and finally stored at -20°C until use.
2.5.5.2 Adhesion and growth on fish mucus
Adhesion ability was evaluated according to Li et al. (2020) with some modifications. Briefly, 100 µl of skin and intestinal mucus was immobilized on a polystyrene microtiter plate by overnight incubation at 4°C. To remove excess mucus, wells were washed twice with 250 μl of PBS. Overnight-grown bacteria (vegetative cells or spores) were centrifuged at 6,000 g for 15 min, washed twice, and resuspended in PBS with fluorescein isothiocyanate (FITC) (100 μg/ml) at 28°C for 1 h in the dark. Labelled bacteria were washed three times with PBS. Then, 100 µl of the labelled bacterial suspension (108 CFU/ml) was added to each well. Plates were incubated at 28°C for 1 h and then washed with PBS to remove unbound bacteria. The bound bacteria were lysed with 200 μl of 1% sodium dodecyl sulphate (SDS) in 0.1 M NaOH at 60°C for 1 h. The fluorescence intensity was measured with the excitation and emission spectrum of 485 nm/530 nm by spectrophotometry. Adhesion was calculated using the formula:
where A is the fluorescence of adhering bacteria and A0 is the fluorescence of initial bacteria.
Moreover, to determine whether the observed bacterial adhesion was due to non-specific adhesion or cell surface hydrophobicity, adhesion assays to bovine serum albumin (BSA) and polystyrene were performed as described above.
The ability of the strain to establish and persist in the gut is essential. For this purpose, the growth of the strain in the intestine and skin mucus was evaluated. In fact, the skin and intestinal mucus were inoculated with the bacterial suspension and spores (106 CFU/mL) and incubated at 28°C for 24 h. Colony-forming units (CFU/mL) were counted. Sterilized mucus-inoculated plates served as controls (Midhun et al., 2017).
2.6 Identification of selected spore formers
The selected spore formers were identified using molecular identification targeting 16S rRNA genes and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) (Ultraflex III, Bruker Daltonics, Bremen, Germany).
Total bacterial DNA was extracted using the InstaGene Matrix resin (Bio-Rad Laboratories Inc., Hercules, CA, USA). PCR amplification was performed using primers 27F and 1492R (Table 1). PCR reaction was prepared as follows: 25 μl of MyTaq Mix (Bioline, London, UK), l μl of each primer (0.7 μmol), and l μl of purified DNA (~50 ng). The samples were amplified under the following conditions: initial denaturation at 97°C for 1 min, followed by 35 cycles of denaturation (95°C for 15 s), annealing (48 for 15 s), and elongation (72°C for 10 s), ending with a final extension step at 72°C for 7 min. The corresponding species identity was obtained by comparative sequence analysis against available sequence data in the National Centre for Biotechnology Information (NCBI) database.
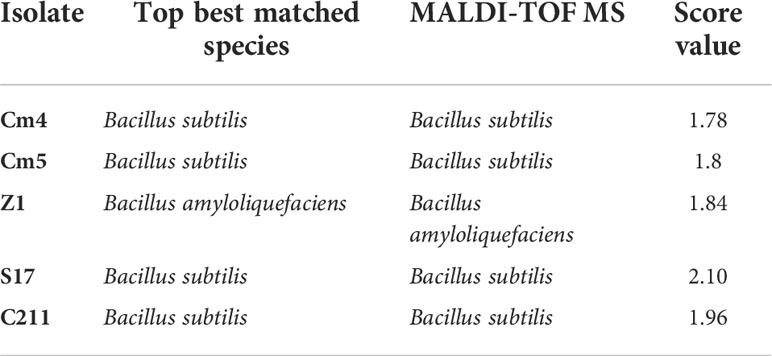
A single colony of selected strains was also analysed by MALDI-TOF MS as described by Altakhis et al. (2021). Bruker Flex Control software was used to run measurements of MALDI-TOF MS which were expressed as score. Based on the report of Bruker software, results of identification were reliable only when the scores generated were ≥1.7 (Han et al., 2021).
2.7 In vitro safety assay
2.7.1 Antibiotic resistance
The antibiotic susceptibility of selected strains was determined by broth microdilution test as described by Araújo et al. (2015). The antibiotics used were ampicillin (0.12–8 μg/ml), vancomycin (0.5–32 μg/ml), gentamicin (2–128 μg/ml), kanamycin (4–256 μg/ml), streptomycin (4–256 μg/ml), erythromycin (0.12–8 μg/ml), clindamycin (0.25–16 μg/ml), tetracycline (0.5–32 μg/ml), and chloramphenicol (1–64 μg/ml). A single colony was suspended in 5 ml of saline solution (0.85% NaCl) to reach an optical density of 0.5 McFarland Scale (108 CFU/ml) and diluted in TSB. Fifty microliters of strain suspension was inoculated with each well containing 50 μl of the different antibiotic concentrations. The inoculated plaques were incubated at 28°C for 18 h; the minimum inhibitory concentrations (MICs) were read as the lowest concentration of antibiotics inhibiting the growth of bacteria and were compared according to EFSA (Muñoz-Atienza et al., 2013). Resistance was defined when the MICs of strains are higher than the respective breakpoint. Staphylococcus aureus CECT794 and Enterococcus faecalis CECT795 were used as controls.
2.7.2 Production of hydrolic enzymes
Haemolysis activity was investigated as previously described by Muñoz-Atienza et al., 2014 (Eaton and Gasson, 2001). Briefly, selected strains previously cultured in TSB with 1.5%NaCl were streaked on horse blood agar plates (bioMérieux, Marcy l’Etoile, France). After 1–2 days at 28°C, the presence of clear zones or green zones of hydrolysis around the colonies revealed β-haemolysis or α-haemolysis, respectively; the absence of clear zones around colonies means γ-haemolysis. Enterococcus faecalis P4 was used as positive control.
Gelatinase production was determined as described by Eaton and Gasson (Eaton and Gasson, 2001). Selected strains grown in TSB broth at 28 C for 16 h were streaked on Todd–Hewitt (Oxoid) agar plates (1.5%, w/v) with 30 g/l gelatin. After overnight incubation, the plates were placed at 4°C for 5 h before examination. The presence of zone of turbidity (protein hydrolysis) around the colonies revealed the activity. Enterococcus faecalis P4 was used as positive control.
The ability of selected strains to hydrolyse primary and secondary bile salts was determined according to Araújo et al. (2015) with some modifications. Briefly, 10 μl of cultures grown in TSB broth was spotted onto TSB agar plates supplemented with 0.5% (w/v) sodium salts of taurocholate or taurodeoxycholate (Sigma-Aldrich, St. Louis, MO, USA) and 0.05% (w/v) L-cysteine (Merck) and incubated at 30°C for 72 h under anaerobic conditions (AnaeroGen, Oxoid). The presence of opaque halos of precipitated deconjugated bile acids around the colonies was considered as a positive result. Fresh faecal slurry of a healthy adult cow was used as the positive control.
2.7.3 PCR detection of potential virulence factors
Detection of genes encoding histidine decarboxylase (hdc), tyrosine decarboxylase (tdc), ornithine decarboxylase (odc), lysine decarboxylase (ldc), surfactin synthetase (sfp), and creolise synthetase (cesA, cesB) was carried out by PCR. Lactobacillus sp. 30a, L. brevis CECT4121, and Bacillus cereus 4086 were used as positive controls. PCR amplifications were performed as previously described by Araújo et al. (2015).
2.8 Statistical analysis
Data were expressed as means ± standard error of the mean (SEM). A statistical analysis of the data was performed using the software package SPSS 19.0. One-way analysis of variance (ANOVA) means and Tukey and Duncan’s tests were applied. Significant differences were accepted to be significant when P< 0.05.
3 Results
3.1 Isolation of spore former strains and screening for antimicrobial activity
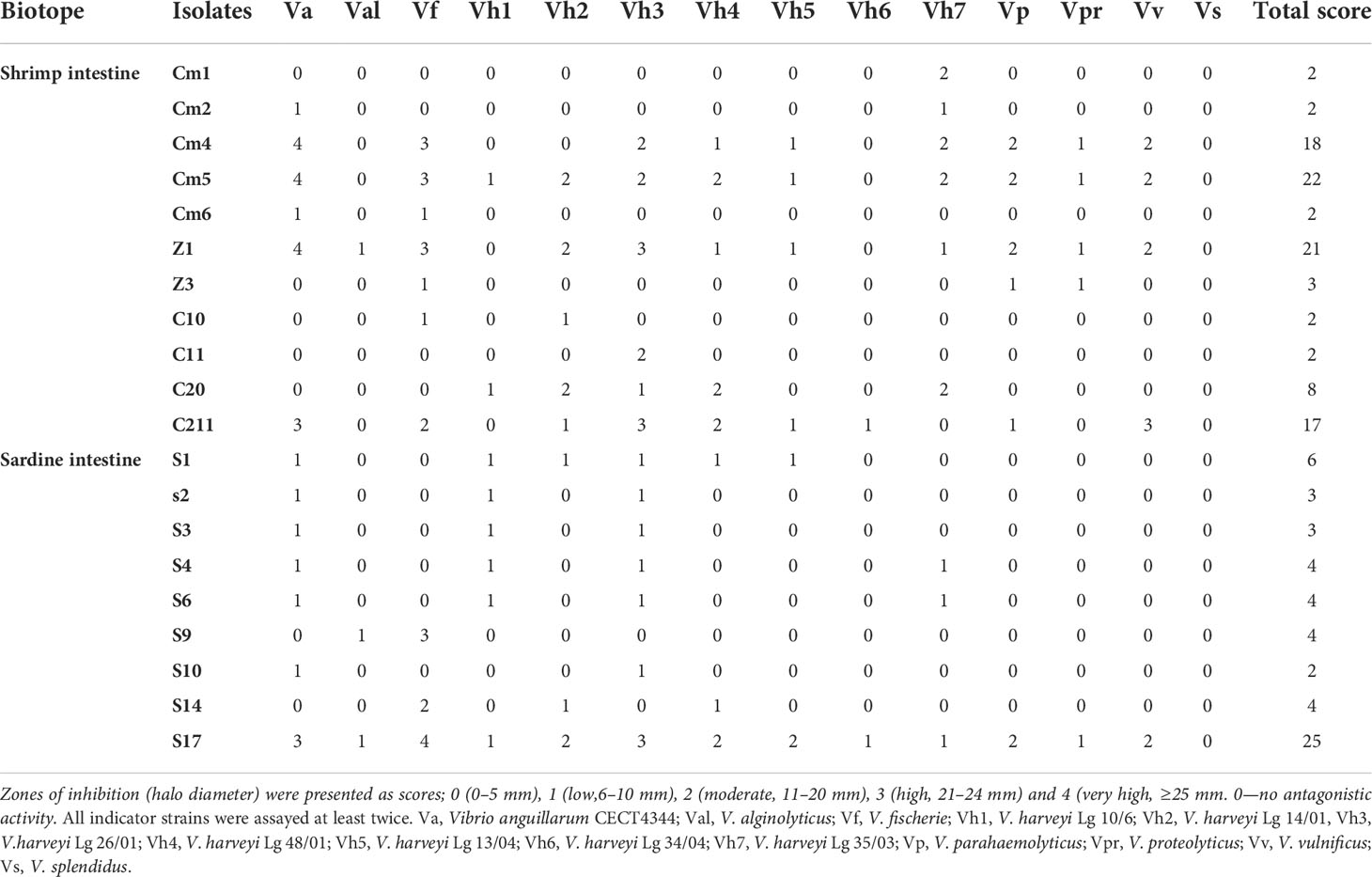
A total of 108 colonies were isolated and purified from sardine and shrimp intestine (47 from Sardina pilchardus and 61 from Penaeus kerathurus). All isolated strains were found to be Gram-positive, catalase positive. Out of 108 isolates, only 20 showed antagonistic activities by the colony overlay method presented as scores (Table 2). Cumulative maximum and minimum scores were 25 and 2, respectively. All 20 exerted direct antimicrobial activity against, at least, two of the indicators (Figure 1). Strain S17 showed a strong antagonistic activity against all tested pathogens.
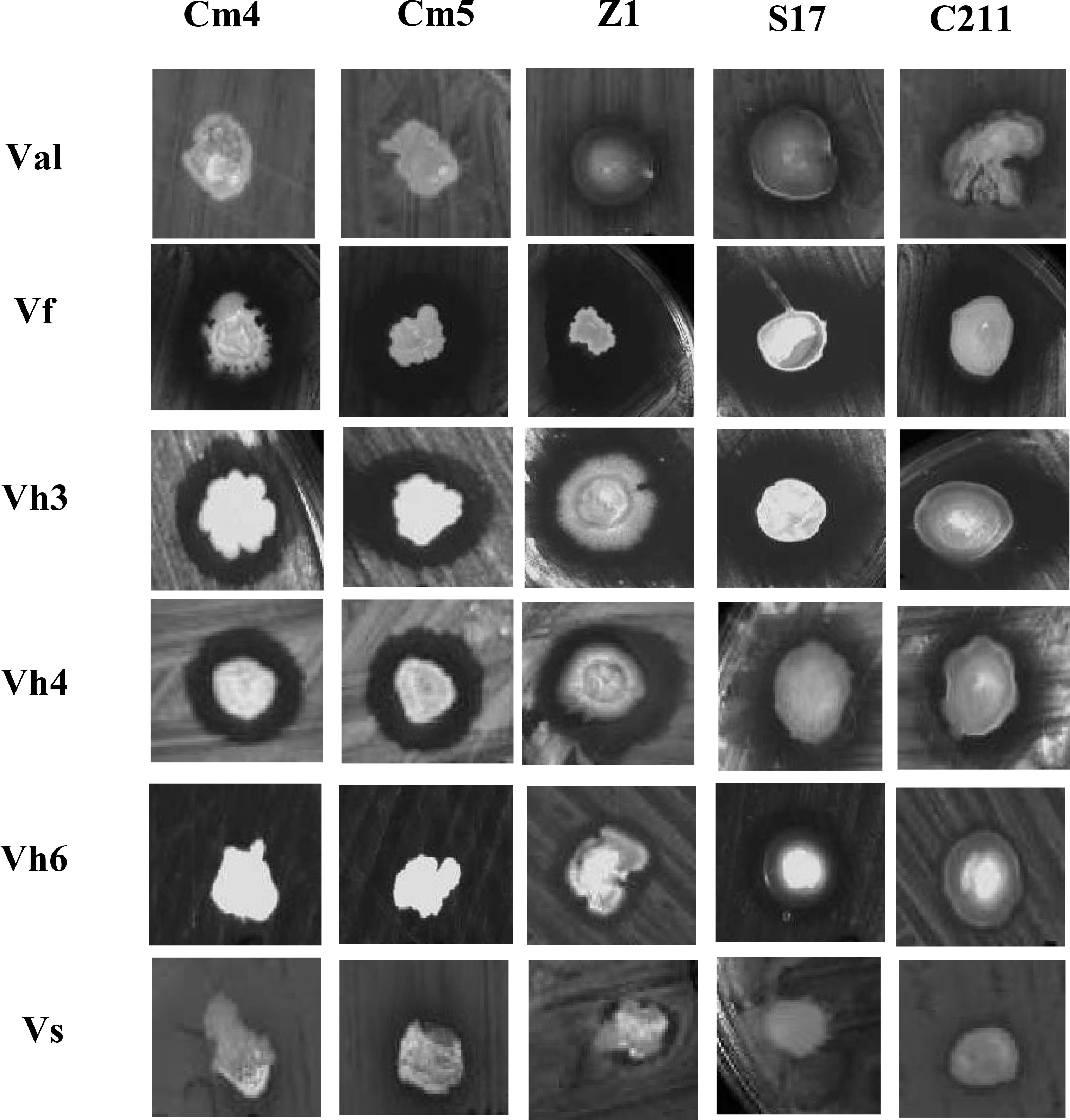
Figure 1 Pictorial overview of antimicrobial activities of selected strains against indicators Vibrios (colony overlay method).Val, V.alginolyticus; Vf, V.fischerie; Vh3, V. harveyi Lg 26/01; Vh4, V. harveyi Lg 48/01; Vh6, V. harveyi Lg 34/04 and Vs, V. spendidus.
Based on the total score, the strong pathogen inhibitory strains (Cm4, Cm5, Z1, S17, and C211) were selected and analysed for extracellular antimicrobial activity assay and for various probiotic potentials.
The inhibition of selected strain CFSs was substantiated by IE values varying in reference time (Figure 2). In majority of hours, inhibition of pathogens was more than 20% for Vibrio anguillarum (Va) and 30% for V. harveyi Lg26/01(Vh3). Moreover, lower inhibition was observed during the initial phase (between 2 and 6 h). On other hand, antibacterial activity of CFS and its nature was as described in Figure 3 and Supplementary Figure S1. All CFSs of selected strains were able to inhibit pathogenic bacteria. The antimicrobial activity of CFS exerted by all these strains withstood after adjustment of pH but disappeared, partially or completely, after heat and proteinase K treatments, thus confirming the proteinaceous nature of the antimicrobial compounds.
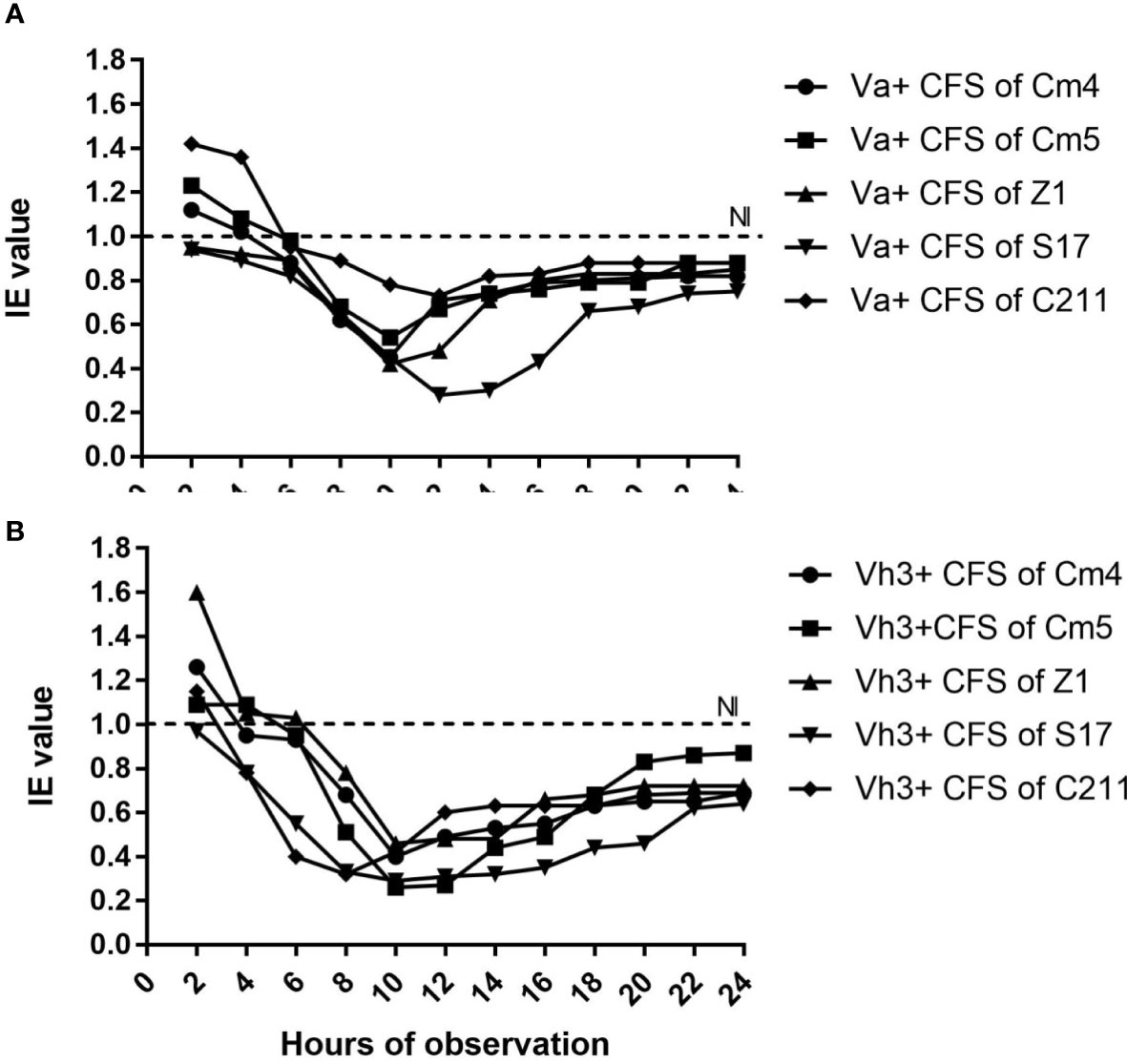
Figure 2 Inhibition efficiency (IE) of selected strains against (A) Vibrio anguillarum CECT4344 (Va) and (B) V.harveyi Lg 26/01 (Vh3) as a function of time. *IE˃1 indicated growth promotion of pathogen, IE=1indicated no inhibition (NI) and IE<1 indicated inhibition of pathogen by CFS of selected strains.
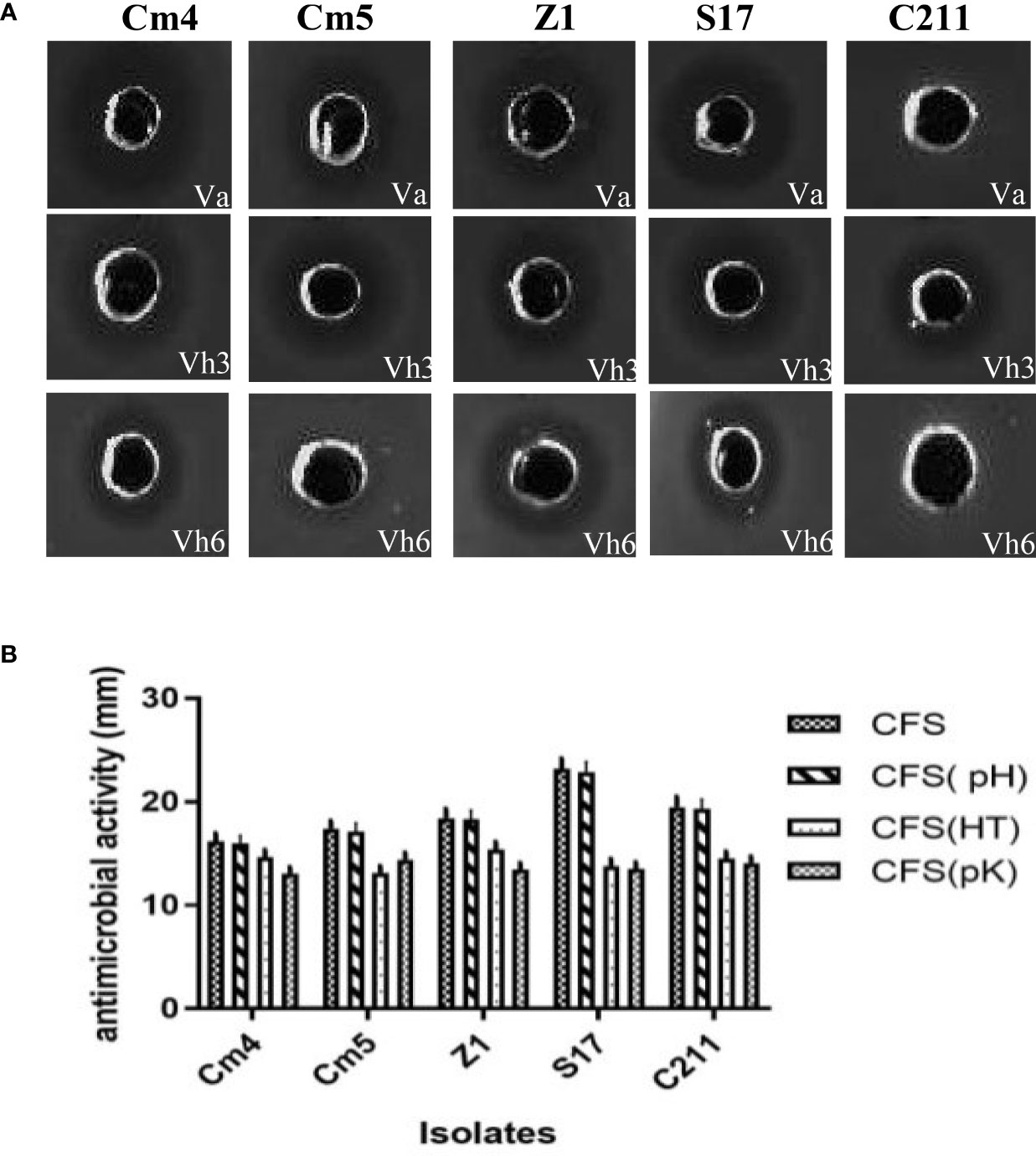
Figure 3 (A) Pictorial overview of antibacterial activity of selected strains against indicators (VA, Vibrio anguillarum CECT4344; Vh3, Vibrio harveyi Lg 26/01and Vh4, V. harveyi Lg 34/04) and (B) nature of their antagonistic activity against Vibrio anguillarum CECT4344 using agar well-diffusion method. *CFS: supernatant without any treatment, CFS (pH): pH neutralization treatment, CFS (HT): heat treatment at 100 ° C, CFS (pK): proteolytic enzyme treatment.
3.2 Sporulation efficiency
All selected strains showed different sporulation efficiencies (Figure 4) and showed spore titres in order of 107–108 spores/ml. The sporulation of S17 and Z1 had the highest rate at 18% and 20% after 24 h and 89% and 92% after 48 h, respectively, while Cm5 had the lowest at 14% and 84% after 24 h and 48 h, respectively.
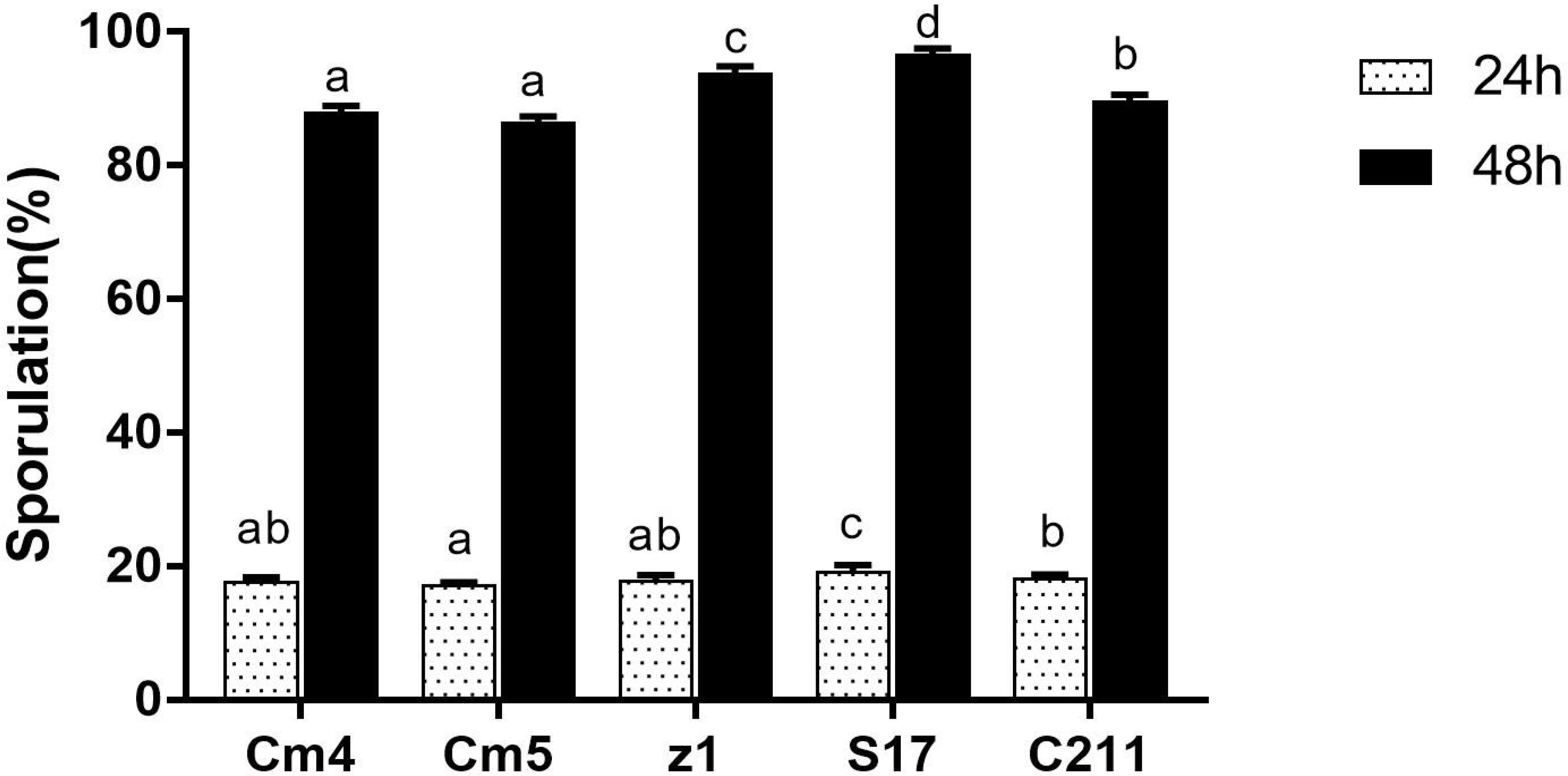
Figure 4 Sporulation efficiency of selected strains. The sporulation is expressed as the percentage of spores relative to vegetative cells after 24 h (white bar) and 48 h (black bar). Data are presented as (mean ± SD, n=3) with different superscript letters denoting a significant differences (p<0.05).
3.3 Probiotic properties of selected spore-forming bacteria
3.3.1 Evaluation of extracellular enzymes and biofilm production
Extracellular enzyme production is wide-ranging among selected strains. The S17 strain exhibited the highest protease and amylase activities. Lipase activity was detected with Cm5, Z1, and C211 (Supplementary Table S1). Further, lipase activity was not detected in sardine isolates. Finally, all the selected strains were able to form biofilm. Among them, S17 had the strongest ability followed by C211 (Figure 5).
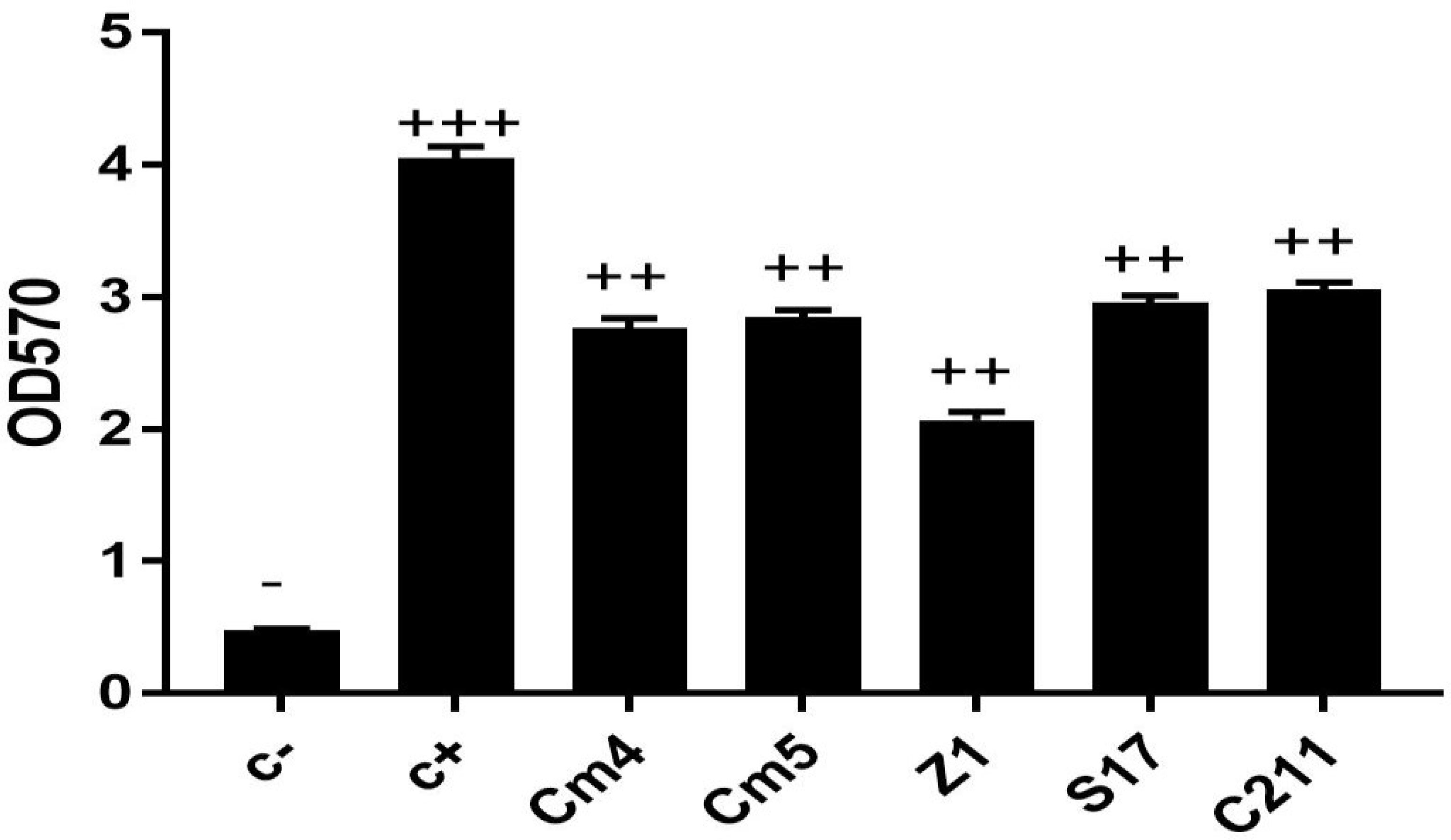
Figure 5 Biofilm production of selected strains. The ability of biofilm production is classified as follows: OD570 < 1, non-biofilm (-); 1< OD570 < 2, weak (+); 2< OD570 < 4, medium (+ +) and OD570>4, strong biofilm (+ + +).
3.3.2 Tolerance to high temperature and pH and bile salt
Vegetative cells and spores were tested for resistance to high temperature, various pH, and bile concentrations.
Selected strains gave promising results after exposure to high temperatures (80°C, 90°C, 100°C for 5 and 10 min). High viability (above 80%) was observed in all selected vegetative cells after exposure to various temperatures compared to control (spore formers without exposure to high temperatures). However, there were no significant differences at different times of exposure among various temperatures for Z1 and S17. At last, the strains Cm4 and C211 had a significant decrease in viability (Supplementary Figure S2).
For the vegetative cells, all selected strains remained viable after 3 h of exposure to pH 2.0 and pH 3.0, but none could tolerate exposure to pH 1.0 (Table 3). Among all tested strains, S17 was found to have the highest resistance to acid conditions, while Cm5 and Z1 showed the lowest acid tolerance. Tested strains also withstood (0.5%–5%) bile concentrations after a 3-h exposure. Thus, S17 showed the highest tolerance to 5% of bile salt, whereas strain CJ3 exhibited the lowest tolerance. However, all spores of tested strains demonstrated significantly high tolerance to gastric and intestinal conditions after a 3-h exposure (Figure 6). Noteworthily, all spores could tolerate exposure to pH 1.0 and remain viable compared to vegetative cells in the same condition.
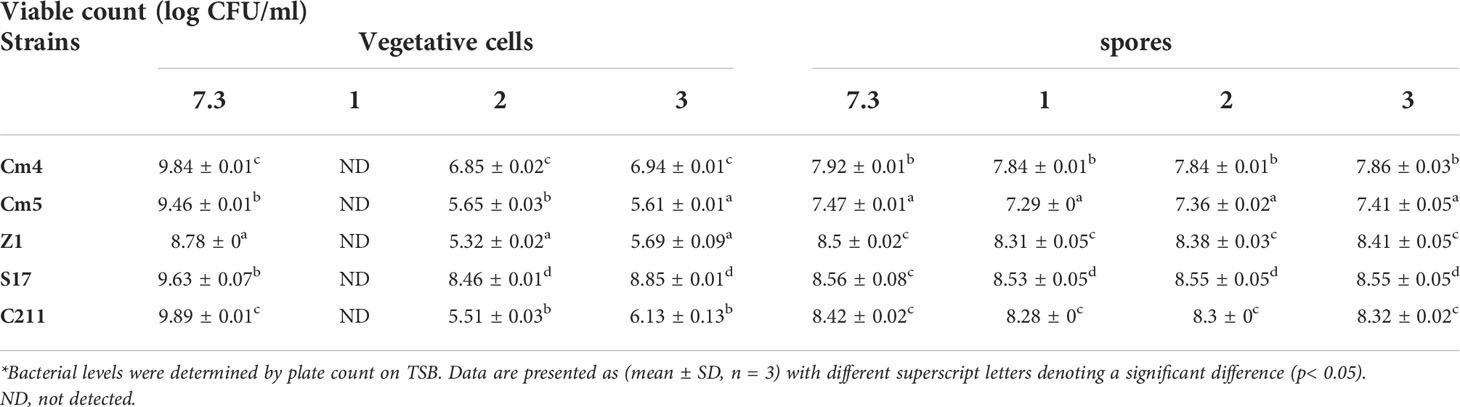
Table 3 Tolerance of vegetative cells and spores at different pH conditions for 3 h at 28°C (log CFU/mL, SD*).
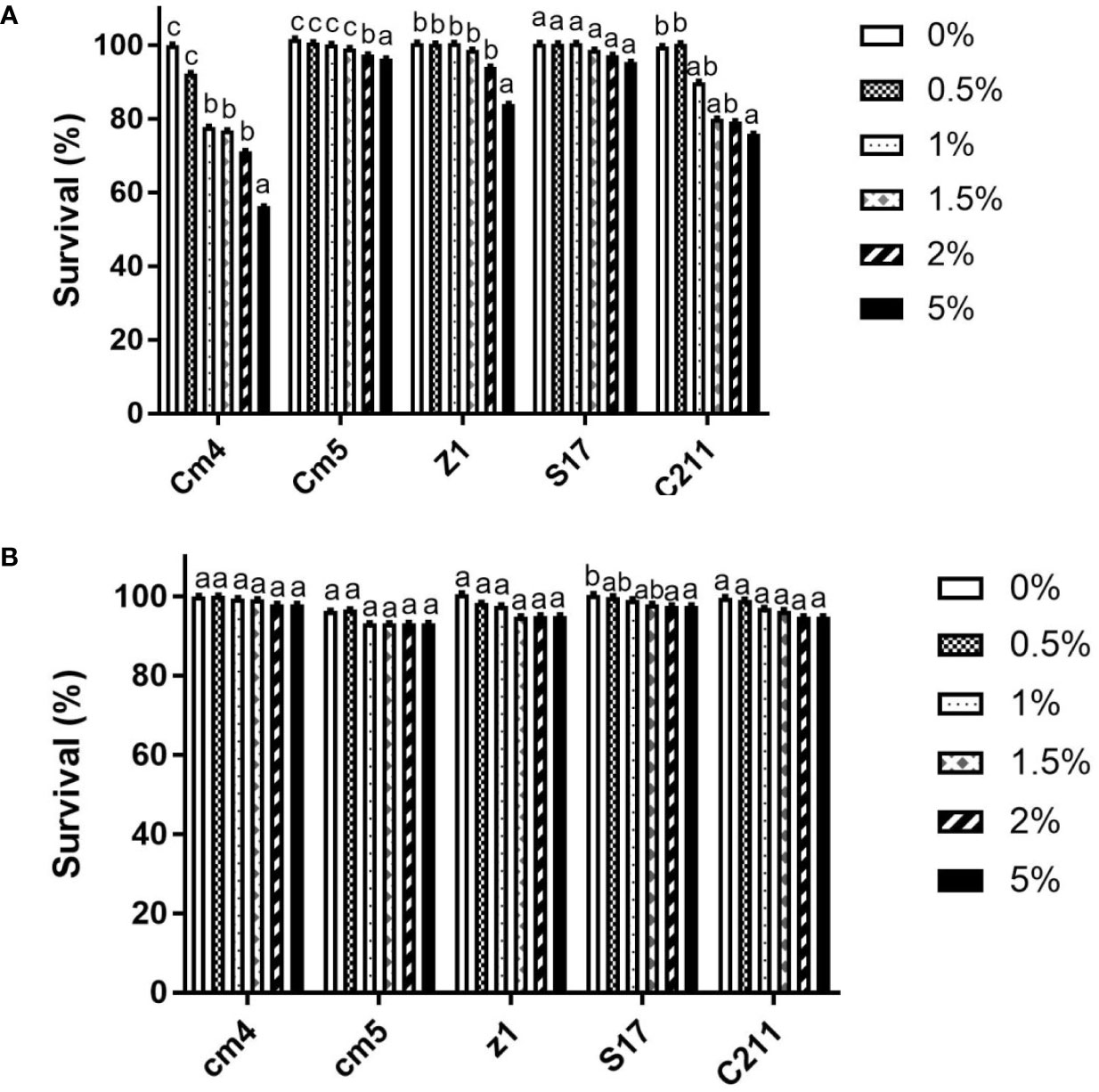
Figure 6 Tolerance of vegetative cells (A) and spores (B) at different bile concentrations for 3h at 28°C (log CFU/mL, SD*). Data are presented as (mean ± SD, n=3) with different superscript letters denoting a significant differences (p<0.05).
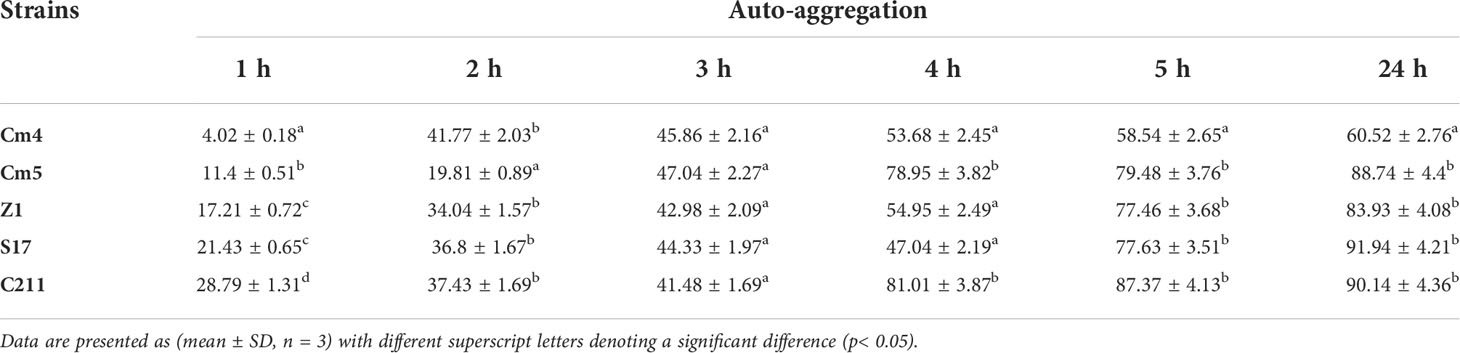
3.3.3 Auto-aggregation and co-aggregation assays
As seen in Table 4, auto-aggregation for selected strains increased with the incubation period and varied from 4% to 92%. All selected strains revealed low auto-aggregation ability (<50%) at the first 3 h. Nonetheless, a significant increase was observed after 24 h. Among them, S17 and C211 exhibited the strongest auto-aggregation ability (92%; 90%, respectively) after 24 h of incubation.
In addition, all tested strains were able to co-aggregate with pathogens significantly, showing co-aggregation percentages above 55% (Figure 7). Among the isolates, S17 showed the highest percentage of co-aggregation against V. fischeri (81%), V. harveyi Lg48/01 (79%), and V. harveyi Lg26/01 (67%) after 6 h of incubation. S17 also presented a strong ability to co-aggregate against V. anguillarum with more than 74%, followed by Cm4 and Cm5.
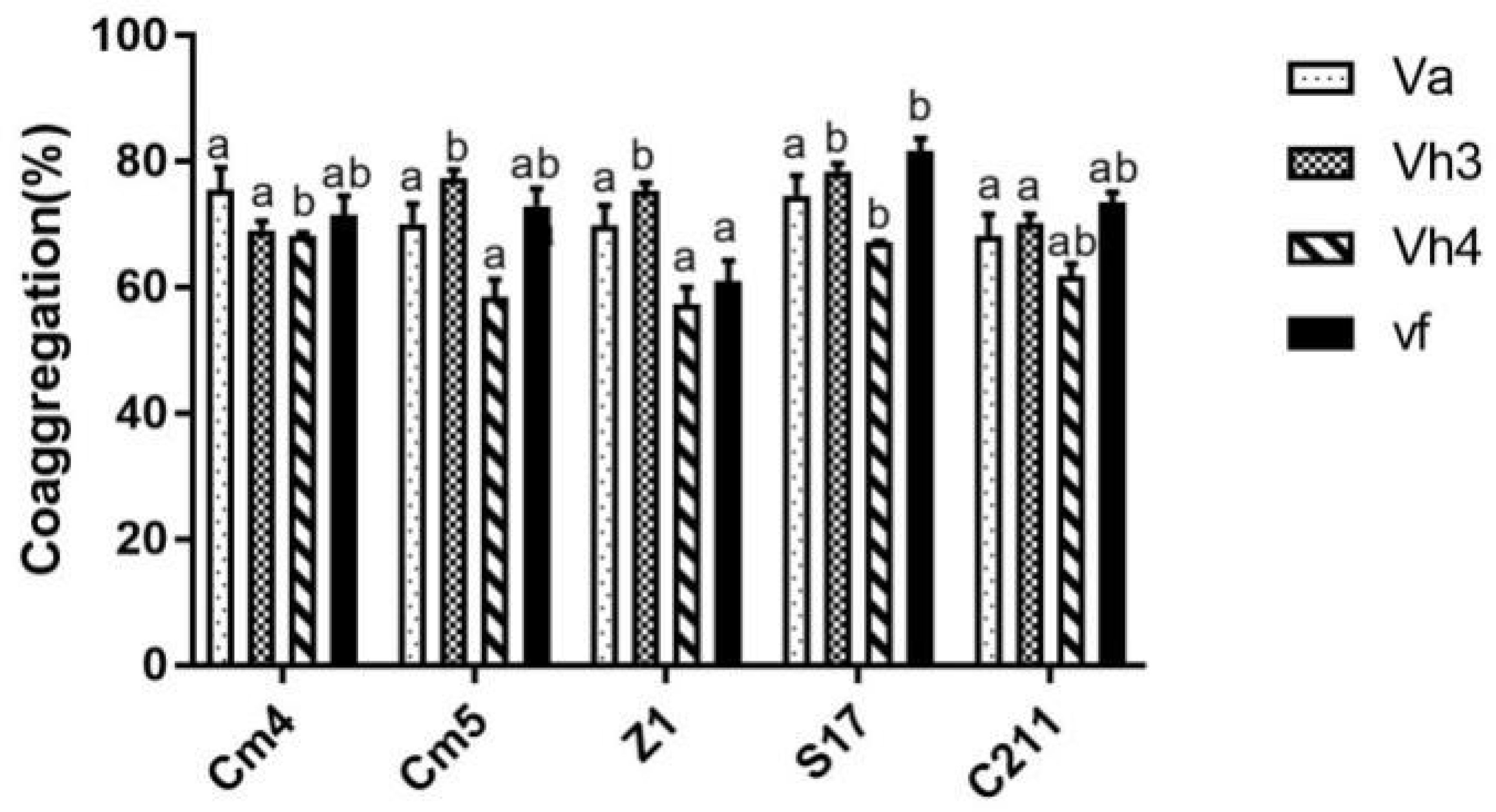
Figure 7 Co-aggregation percentages of selected strains with pathogens after 6h.Va, Vibrio anguillarum CECT4344; Vh3, V. harveyi Lg 26/01; Vh4, V. harveyi Lg 48/01; Vf, V. fischerie. Data are presented as mean ± SD (n=3) with different superscript letters denoting a significant differences (p<0.05).
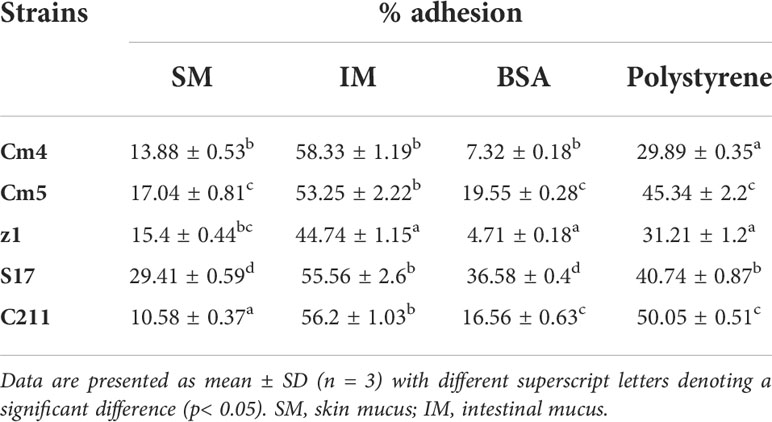
3.3.4 Adhesion and growth on fish mucus
The adhesion ability of the tested strains (vegetative cells) to mucus is shown in Table 5. All tested strains were highly capable of binding to intestinal mucus (44.7 to 58.3%) but less to skin mucus (10.6% to 29.4%). S17 showed the highest, both skin and intestinal mucus adhesion with more than 29% and 58%, respectively. The adhesion ability of spores was found to be lower than that of its vegetative cells for skin and intestinal mucus (Figure 8A). All the spores were more able for adhesion to intestinal mucus than skin mucus, ranging from 8% to 60%. All tested strains (vegetative cells and spores) adhered significantly less to BSA (5% to 37%) and polystyrene (19% to 50%) compared to skin and intestinal mucus.
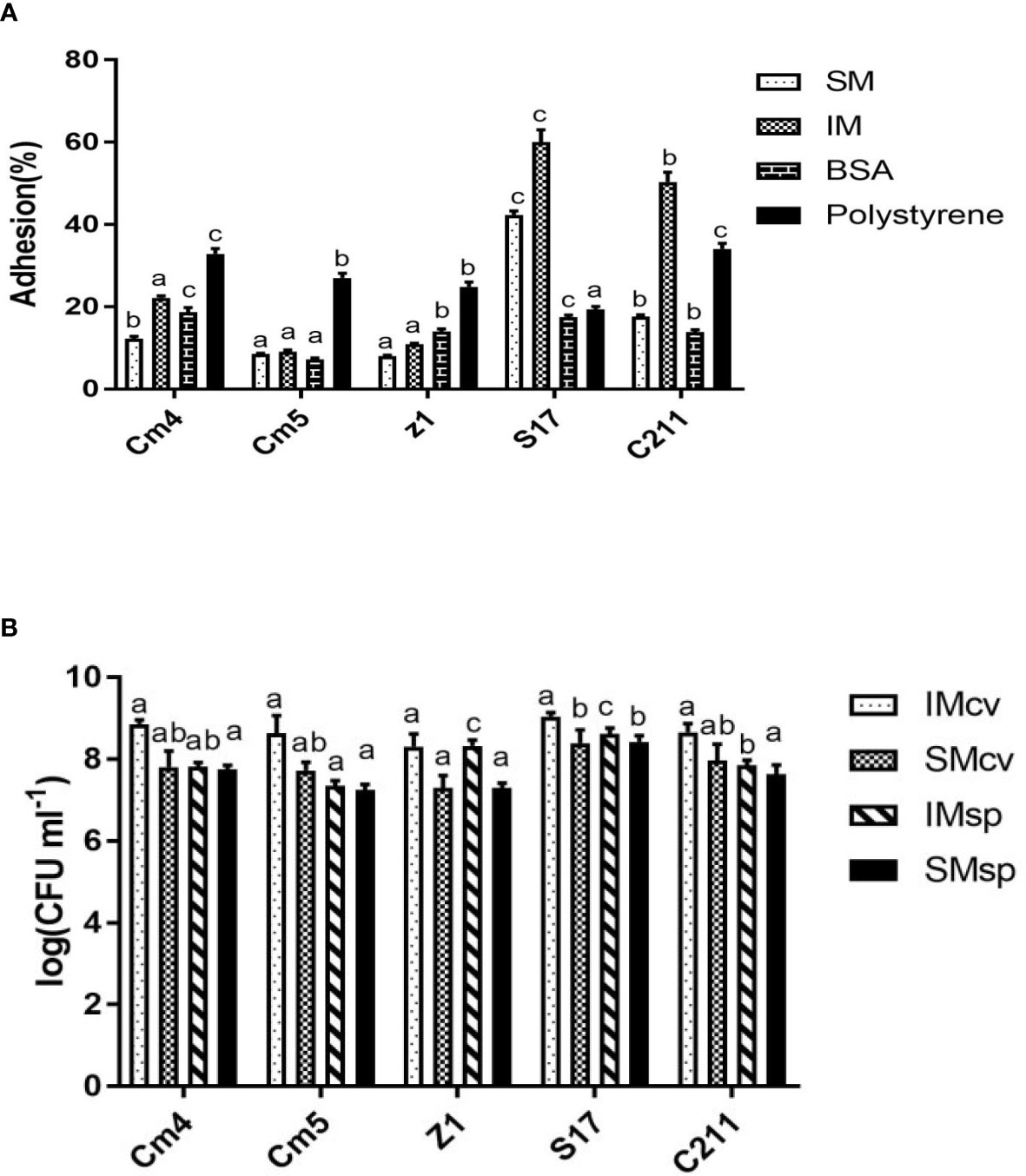
Figure 8 (A) Adhesions of spores to fish mucus, BSA and polystyrene and (B) Growth of selected strains on intestinal and skin mucus of Sparus aurata. Data are presented as mean ± SD (n=3) with different superscript letters denoting a significant differences (p<0.05). * IM, intestinal mucus; SM, skin mucus; cv, vegetative cells; sp, spores.
All tested strains (vegetative cells and spores) were able to grow in fish mucus collected from the skin and intestine of S. aurata (Figure 8B). The growth of vegetative cells was found to be higher than that of their spores for skin and intestinal mucus. Interestingly, growth of vegetative cells on intestinal mucus was observed to be higher compared to growth on skin mucus. The significantly highest growth was observed for S17, for both skin and intestinal mucus, respectively.
3.4 Identification and in vitro safety assay
The selected strains were subjected to partial 16S rDNA gene sequencing and MALDI-TOF MS. According to these methods, all selected strains were identified as Bacillus spp. The isolates were identified as B. subtilis (Cm4, Cm5, S17, and C211) and B. amyloliquefaciens (Z1) by MALDI-TOF MS (Table 6) which was confirmed by PCR. In addition, all selected spore formers were found to be susceptible to tested antibiotics.
All strains exhibited haemolysis on blood agar (β-haemolytic) except S17, which was negative for haemolytic activity (γ-haemolytic) (Supplementary Table 2). More analysis revealed that Z1 and C211 were found to exhibit bile salt deconjugation, and all selected isolates showed gelatinase production except S17 (Supplementary Table S2). Furthermore, PCR screening for the presence of biogenic amines and toxins showed that none of the tested strains encoded any of the virulence genes searched hdc, tdc, odc, sfp, cesA, or cesB, with the exception of ldc which was detected in Cm4 and C211 (Supplementary Table 2).
4. Discussion
During the past decades, aquaculture production has faced many threats due to the outbreak of diseases, i.e., Vibriosis (Yılmaz et al., 2021). In order to control or prevent fish pathogens, probiotics are used as a viable and friendly alternative (Balcázar et al., 2008; Kavitha et al., 2018; Khan et al., 2021). Gastrointestinal microbiota is considered as the best source to isolate and select potential probiotics. Indeed, fish and shrimp intestine constitute a dynamic ecosystem containing diverse unexplored microorganisms and playing a crucial role in animal health (Romero et al., 2014; Zhou et al., 2014; Kuebutornye et al., 2020; Medina et al., 2020). Numerous studies assumed that probiotics from target marine animals and from the same ecological niche of pathogens would be more effective (Midhun et al., 2017; Kuebutornye et al., 2020; Yamashita et al., 2020; Santos et al., 2021). In the present investigation, spore-former strains were isolated from sardine and shrimp intestine and were evaluated for their probiotic potential and their safety to select the most putative as probiotic for aquaculture.
An initial prescreening was carried out based on antimicrobial activity against Vibriosis which is one of major pathogens in aquaculture. From 108 isolates, only 20 showed antagonistic activity by the colony overlay method against, at least, two of tested Vibriosis such as V. harveyi, V. anguillarum, and V. fischeri. Previous studies indicated that Bacillus species, including B. pumilus (Lei et al., 2015), B. amyloliquefaciens (Cai et al., 2019; Medina et al., 2020), and B. subtilis (Zhou et al., 2019), exhibited antagonistic activity against Vibriosis (Amoah et al., 2021). Similar studies also reported that some Bacillus inhibit the growth of various Vibrio species (V. harveyi, V. alginolyticus, V. vulnificus, and V. parahaemolyticus) (Prieto et al., 2014; Meidong et al., 2018; Li et al., 2019; Zhou et al., 2019). Later on, selected strains with promising direct antimicrobial activity were tested for extracellular inhibitory activity by ADT and microtiters. In this respect, only five strains were found to produce proteinaceous compounds and B. subtilis S17 showed the strongest antagonistic activity against all tested Vibrio species. In fact, the antibacterial activity of selected strains is probably explained by the production of inhibitory components such as organic acids, hydrogen peroxide, and bacteriocins or by competition for adhesion sites and/or nutrients in the intestine (Pinto et al., 2020; Santos et al., 2021). Hence, published reports revealed that the antibacterial substances not only inhibit pathogen’s growth but also increase the resistance of the host against pathogens (Verschuere et al., 2000; Guo et al., 2016; Midhun et al., 2017; Samson et al., 2020; Khan et al., 2021).
Unlike lactic acid bacteria, Bacillus sp. is considered to be a very stable strain due to its sporulation ability. Their endospores are resistant to harsh conditions, including high temperature, UV and acidity, drought, freezing, radiation, and rising oxygen levels. Relatively, Bacillus spores are able to survive throughout the simulation of GIT (Barbosa et al., 2005; Kuebutornye et al., 2020). In this present work, the sporulation rates of selected strains were more than 80% after 48 h of incubation. However, the differences in sporulation rates observed among the bacilli tested may be due to a different mechanism of sporulation process (Barbosa et al., 2005; Prieto et al., 2014; Banerjee and Ray, 2017).
Probiotics not only reduce the risk of fish infection in aquaculture by inhibiting the growth of pathogens but also could promote animal health. Several studies report that Bacillus sp. is able to produce digestive enzymes, including amylase, protease, lipase, cellulose, and xylanase (Hmani et al., 2017; Midhun et al., 2017). The present study confirms the ability of selected bacterial strains to produce extracellular enzymes including amylase, protease, and lipase. Indeed, these enzymes facilitate the digestion of fish diet and can help and improve the growth rate. Therefore, application of probiotics capable of producing enzymes is gaining attention to promote a nutritional benefit in cultivable fish (Benhamed et al., 2014; Sharifuzzaman et al., 2018).
Moreover, biofilm is a complex and heterogenic aggregation of microorganisms. The probiotic biofilm has many benefits to host intestinal health including increasing the gut resistance by producing secondary metabolites, increasing the colonization efficiency, protecting the gut epithelium by competing with pathogenic strains for adherence sites, and inducing immune reactions to the gut pathogens in the host (Midhun et al., 2017; Banerjee and Ray, 2017; Midhun et al., 2017; Chauhan and Singh, 2019). In this study, all selected strains were capable of forming biofilm at high levels. Among them, strains S17 and C211 had the strongest ability to form biofilm.
During the production of animal feed production, heat treatment is considered as an essential process to get rid of vegetative cells (Banerjee and Ray, 2017; Kuebutornye et al., 2020). In this report, all tested strains showed higher survivability percentages above 80% during heat treatment (80°C, 90°C, 100°C) which were mentioned on previous findings (Kuebutornye et al., 2020; Amoah et al., 2021). Furthermore, probiotics are intended to colonize the GIT of fish and to restore the balance of intestinal microflora. The candidate strains should tolerate GIT conditions. In fact, ingested probiotic strains need to survive the acidity of stomach and fish bile in the small intestine. Moreover, bile plays an important role in the mechanism (specific and non-specific) of defence in the gut. Hence, probiotic strains are required to tolerate bile salts which are so toxic for bacteria due to their effect on the structure of the membrane (Ramesh et al., 2015; Egerton et al., 2018; Feng et al., 2019; Samson et al., 2020). In the present study, the vegetative cells and spores of selected strains were tested in GIT conditions. No vegetative cells could tolerate pH 1.0 while the majority of tested cells withstand a low pH above 1.0 and a wide range of bile concentrations up to 5%. Previous studies have reported the ability of some strains of Bacillus to survive the stomach and intestinal environment where they could work effectively (Shinde et al., 2019; Medina et al., 2020), whereas a small reduction of spore count of tested strains was observed and could be due to their germination (Barbosa et al., 2005; Sahoo et al., 2015). This finding suggests that those can be used as probiotic as they could tolerate the harsh gastric acid and toxic bile.
Determination of bacterial cell surface properties, i.e., hydrophobicity, auto-aggregation, and co-aggregation, is an indirect method for the adhesion ability of probiotic strains. Further, aggregating probiotic bacteria may help not only adhesion and colonization of the GIT but also the modulation of the immune system (Balakrishna, 2013; Dutta et al., 2018; Kaktcham et al., 2018; Khan et al., 2021). In the current study, all tested strains showed auto-aggregation ability ranging from 4% to 92% after 24 h of incubation. Among them, S17 and C211 exhibited high auto-aggregation ability (92%; 90%, respectively), in accordance with previous findings (Meidong et al., 2017; Khan et al., 2021; Amoah et al., 2021). Moreover, a co-aggregation assay was developed to quantify inter-bacterial adherence (Dutta et al., 2018; Rungsirivanich et al., 2020). Co-aggregation capacity is considered as a way to exclude pathogens from the host. According to different investigations, co-aggregation ability can increase the competition of an epithelial cell’s receptor and may protect GIT against undesired microorganisms (Balakrishna, 2013; Dutta et al., 2018; Kaktcham et al., 2018; Meidong et al., 2018; Rungsirivanich et al., 2020). In the present study, all strains tested were able to co-aggregate with pathogens, showing inhibition percentages above 55%. The result also points out that strain S17 showed the highest percentage of co-aggregation against Vibrio fischeri (81%), V. harveyi Lg48/01 (79%), and V. harveyi Lg26/01 (67%) after 6 h of incubation. Dutta et al. found that B. subtilis LR3H1A and B. tequilensis LR3F3P showed a strong co-aggregation with several pathogens (Khan et al., 2021).
The adhesion of probiotic strains to mucus is required and is considered as an important criterion for probiotic selection. Adherence to GIT is the first step in probiotic strains for colonization, stimulation of the immune system, and antagonistic activity. Upon ingestion in GIT, probiotic strains have to attach to the border of microvilli or to the mucus to prevent the colon sweep. Cell adhesion is a complex process which involves contact between the membrane of bacterial cell and surface (Mbozo et al., 2017; Dutta et al., 2018; Rungsirivanich et al., 2020; Gutiérrez Falcón et al., 2021). In fact, many factors including electrostatic interactions, hydrophobicity, steric forces, components of the surface, pH of the milieu, and viscosity are involved (Balcázar et al., 2008; Muñoz-Atienza et al., 2014; Chauhan and Singh, 2019). In this study, all tested strains (vegetative cells and spores) were highly capable of binding to the intestinal mucus but less to skin mucus. This higher adhesion capacity may be due to the presence of specific receptors on the intestinal mucus or may be related to mucus composition (Balcázar et al., 2008). However, all tested Bacillus adhered less to polystyrene and BSA compared to intestinal mucus. This binding may involve specific interactions. Similar results were reported by Balcázar et al. and Muñoz-Atienza et al. (Balcázar et al., 2008; Muñoz-Atienza et al., 2014). Once probiotic strains adhered, the next step involved their mucus growth in order to establish in the intestine of marine animals (Mbozo et al., 2017). In the present investigation, all vegetative cells and spores of tested strains grew well in the skin and intestine mucus of S. aurata, which indicates that our probiotic strains will be able to bind, establish, and remain in the intestine.
Safety assessment is an important precondition for probiotic evaluation. Before the use of probiotics, it is mandatory to confirm that no pathogenic and harmful effects may occur in marine hosts (Verschuere et al., 2000; Medina et al., 2020; Pinto et al., 2020). In the current investigation, all selected stains were found to be susceptible to tested antibiotics demanded by EFSA. This result may ensure the inability and scarcity to transfer antibiotic resistance genes to gut bacteria, thus assuring their safety for aquaculture application (Gutiérrez Falcón et al., 2021). Regarding hydrolic enzymes and virulence factors, S17 did not show haemolytic activity (γ-haemolytic), gelatinase production, and bile salt deconjugation. In addition, S17 appeared to be free of studied virulence factors. Our results are in accordance with the previous study (Hsieh et al., 2004; Shinde et al., 2019). With this regard, candidate probiotic S17 may be considered as a safe strain.
5 Conclusions
In the present study, 108 spore-former strains were isolated from sardine and shrimp intestine. Among all tested strains, only B. subtilis S17 possesses suitable probiotic potential for Vibriosis control based on antimicrobial activity, heat and GIT tolerances, extracellular enzyme production, adhesion ability, and safety assessment. Afterward, in vivo evaluation studies are required to determine its real-life applications.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MZ378829.
Author contributions
MJ, IA and MamBA designed the study. MJ conducted the study, analysed the data, and wrote the manuscript. AH, WB, and ManBA participated in its organization and helped to draft the manuscript. MJ and MamBA revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a grant from Astrum biotech the Ministry for Higher Education and Scientific Research, Tunisia.
Acknowledgments
The authors would like to thank Miguel A. Morinigo (Department of Microbiology, Faculty of Science University of Malaga, Spain) for providing us all the Vibrio strains used in this study. The authors are grateful to Professor Luis M.Cintas Izarra (Departmental of Nutrition and Food Science Veterinary Faculty University of Complutense Madrid, Spain) who provided us control strains used in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.884244/full#supplementary-material
References
Alonso S., Castro M. C., Berdasco M., de la Banda I. G., Moreno-Ventas X., de Rojas A. H. (2019). Isolation and partial characterization of lactic acid bacteria from the gut microbiota of marine fishes for potential application as probiotics in aquaculture. Probiotics. antimicrob. proteins. 11 (2), 569–579. doi: 10.1007/s12602-018-9439-2
Altakhis M., Pillidge C. J., Osborn A. M., Torley P. J., Kaur M. (2021). Assessment of the potential use of MALDI-TOF MS for the identification of bacteria associated with chilled vacuum-packaged lamb meat. Meat. Science. 177, 108508. doi: 10.1016/j.meatsci.2021.108508
Amoah K., X-h D., Tan B.-p., Zhang S., Kuebutornye F. K. A., Chi S.-y., et al. (2021). In vitro assessment of the safety and potential probiotic characteristics of three bacillus strains isolated from the intestine of hybrid grouper (Epinephelus fuscoguttatus♀× e. lanceolatus♂). Front. Vet. Science. 8. doi: 10.3389/fvets.2021.675962
Araújo C., Muñoz-Atienza E., Nahuelquín Y., Poeta P., Igrejas G., Hernández P. E., et al. (2015). Inhibition of fish pathogens by the microbiota from rainbow trout (Oncorhynchus mykiss, walbaum) and rearing environment. Anaerobe. 32, 7–14. doi: 10.1016/j.anaerobe.2014.11.001
Balakrishna A. (2013). In vitro evaluation of adhesion and aggregation abilities of four potential probiotic strains isolated from guppy (Poecilia reticulata). Braz. Arch. Biol. Technol. 56 (5), 793–800. doi: 10.1590/S1516-89132013000500010
Balcázar J. L., De Blas I., Ruiz-Zarzuela I., Cunningham D., Vendrell D., Muzquiz J. L. (2006). The role of probiotics in aquaculture. Vet. Microbiol. 114 (3-4), 173–186. doi: 10.1016/j.vetmic.2006.01.009
Balcazar J. L., De Blas I., Ruiz-Zarzuela I., Vendrell D., Girones O., Muzquiz J. L. (2007). Sequencing of variable regions of the 16S rRNA gene for identification of lactic acid bacteria isolated from the intestinal microbiota of healthy salmonids. Comp. immunol. Microbiol. Infect. diseases. 30 (2), 111–118. doi: 10.1016/j.cimid.2006.12.001
Balcázar J. L., Vendrell D., de Blas I., Ruiz-Zarzuela I., Muzquiz J. L., Girones O. (2008). Characterization of probiotic properties of lactic acid bacteria isolated from intestinal microbiota of fish. Aquaculture 278 (1-4), 188–191. doi: 10.1016/j.aquaculture.2008.03.014
Banerjee G., Ray A. K. (2017). The advancement of probiotics research and its application in fish farming industries. Res. Vet. Sci. 115, 66–77. doi: 10.1016/j.rvsc.2017.01.016
Barbosa T. M., Serra C. R., La Ragione R. M., Woodward M. J., Henriques A. O. (2005). Screening for bacillus isolates in the broiler gastrointestinal tract. Appl. Environ. Microbiol. 71 (2), 968–978. doi: 10.1128/AEM.71.2.968-978.2005
Beltrán J. M. G., Espinosa C., Guardiola F. A., Esteban MÁ. (2017). Dietary dehydrated lemon peel improves the immune but not the antioxidant status of gilthead seabream (Sparus aurata l.). Fish. shellfish. Immunol. 64, 426–436. doi: 10.1016/j.fsi.2017.03.042
Benhamed S., Guardiola F. A., Mars M., Esteban MÁ. (2014). Pathogen bacteria adhesion to skin mucus of fishes. Vet. Microbiol. 171 (1-2), 1–12. doi: 10.1016/j.vetmic.2014.03.008
Cai Y., Yuan W., Wang S., Guo W., Li A., Wu Y., et al. (2019). In vitro screening of putative probiotics and their dual beneficial effects: To white shrimp (Litopenaeus vannamei) post larvae and to the rearing water. Aquaculture. 498, 61–71. doi: 10.1016/j.aquaculture.2018.08.024
Cao L., Pan L., Gong L., Yang Y., He H., Li Y., et al. (2019). Interaction of a novel bacillus velezensis (BvL03) against aeromonas hydrophila in vitro and in vivo in grass carp. Appl. Microbiol. Biotechnol. 103 (21-22), 8987–8999. doi: 10.1007/s00253-019-10096-7
Caruffo M., Navarrete N., Salgado O., Díaz A., López P., García K., et al. (2015). Potential probiotic yeasts isolated from the fish gut protect zebrafish (Danio rerio) from a vibrio anguillarum challenge. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01093
Chauhan A., Singh R. (2019). Probiotics in aquaculture: a promising emerging alternative approach. Symbiosis. 77 (2), 99–113. doi: 10.1007/s13199-018-0580-1
Cintas L. M., Rodriguez J. M., Fernandez M. F., Sletten K., Nes I. F., Hernandez P. E., et al. (1995). Isolation and characterization of pediocin L50, a new bacteriocin from pediococcus acidilactici with a broad inhibitory spectrum. Appl. Environ. Microbiol. 61 (7), 2643–2648. doi: 10.1128/aem.61.7.2643-2648.1995
Daniels N. A., Shafaie A. (2000). A review of pathogenic vibrio infections for clinicians. Infections Med. 17 (10), 665–685.
do Vale Pereira G., da Cunha D., Pedreira Mourino J., Rodiles A., Jaramillo-Torres A., Merrifield D. (2017). Characterization of microbiota in arapaima gigas intestine and isolation of potential probiotic bacteria. J. Appl. Microbiol. 123 (5), 1298–1311. doi: 10.1111/jam.13572
Dutta D., Banerjee S., Mukherjee A., Ghosh K. (2018). Potential gut adherent probiotic bacteria isolated from rohu, labeo rohita (Actinopterygil : Cypriniformes:Cyprinidae): Characterisation, exo-enzyme production, pathogen inhibition, cell surface hydrophobicity, and biofilm formation. Acta Ichthyologica Piscatoria. 48 (3), 221–233. doi: 10.3750/AIEP/02251
Eaton T. J., Gasson M. J. (2001). Molecular screening of enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67 (4), 1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001
Egerton S., Culloty S., Whooley J., Stanton C., Ross R. P. (2018). The gut microbiota of marine fish. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00873
Ehling-Schulz M., Fricker M., Scherer S. (2004). Identification of emetic toxin producing bacillus cereus strains by a novel molecular assay. FEMS Microbiol. letters. 232 (2), 189–195. doi: 10.1016/S0378-1097(04)00066-7
Emam A. M., Dunlap C. A. (2020). Genomic and phenotypic characterization of bacillus velezensis AMB-y1; a potential probiotic to control pathogens in aquaculture. Antonie van Leeuwenhoek 113 (12), 2041–2052. doi: 10.1007/s10482-020-01476-5
FAO/WHO/OIE (2006). Expert consultation on antimicrobial use in aquaculture and antimicrobial resistance.
Feng P., Ye Z., Kakade A., Virk A., Li X., Liu P. (2019). A review on gut remediation of selected environmental contaminants: Possible roles of probiotics and gut microbiota. Nutrients. 11 (1), 22. doi: 10.3390/nu11010022
Fernandes S., Kerkar S. (2019). Bacterial probiotics over antibiotics: A boon to aquaculture. Adv. Biol. Sci. Res. Elsevier;, 2021 (13), 1572–1584. doi: 10.1016/B978-0-12-817497-5.00014-8
Fuller R. (1989). Probiotics in man and animals. J. Appl. bacteriol. 66 (5), 365–378. doi: 10.1111/j.1365-2672.1989.tb05105.x
Ghanei-Motlagh R., Mohammadian T., Gharibi D., Menanteau-Ledouble S., Mahmoudi E., Khosravi M., et al. (2020). Quorum quenching properties and probiotic potentials of intestinal associated bacteria in Asian Sea bass lates calcarifer. Mar. Drugs 18 (1), 23. doi: 10.3390/md18010023
Guo X., Chen D.-D., Peng K.-S., Cui Z.-W., Zhang X.-J., Li S., et al. (2016). Identification and characterization of bacillus subtilis from grass carp (Ctenopharynodon idellus) for use as probiotic additives in aquatic feed. Fish. Shellfish. Immunol. 52, 74–84. doi: 10.1016/j.fsi.2016.03.017
Gutiérrez Falcón A., Padilla D., Real F., Ramos Sosa M. J., Acosta-Hernández B., Sánchez Henao A., et al. (2021). Screening of new potential probiotics strains against photobacterium damselae subsp. piscicida for marine aquaculture. Animals. 11 (7), 2029. doi: 10.3390/ani11072029
Han S.-S., Jeong Y.-S., Choi S.-K. (2021). Current scenario and challenges in the direct identification of microorganisms using MALDI TOF MS. Microorganisms. 9 (9), 1917. doi: 10.3390/microorganisms9091917
Hmani H., Daoud L., Jlidi M., Jalleli K., Ali M. B., Brahim A. H., et al. (2017). Ali MB. a bacillus subtilis strain as probiotic in poultry: selection based on in vitro functional properties and enzymatic potentialities. J. Ind. Microbiol. Biotechnol. 44 (8), 1157–1166. doi: 10.1007/s10295-017-1944-x
Horwood P. F., Burgess G. W., Jane Oakey H. (2004). Evidence for non-ribosomal peptide synthetase production of cereulide (the emetic toxin) in bacillus cereus. FEMS Microbiol. letters. 236 (2), 319–324. doi: 10.1111/j.1574-6968.2004.tb09664.x
Hsieh F.-C., Li M.-C., Lin T.-C., Kao S.-S. (2004). Rapid detection and characterization of surfactin-producing bacillus subtilis and closely related species based on PCR. Curr. Microbiol. 49 (3), 186–191. doi: 10.1007/s00284-004-4314-7
Kaktcham P. M., Temgoua J.-B., Zambou F. N., Diaz-Ruiz G., Wacher C., de Lourdes Pérez-Chabela M. (2018). In vitro evaluation of the probiotic and safety properties of bacteriocinogenic and non-bacteriocinogenic lactic acid bacteria from the intestines of Nile tilapia and common carp for their use as probiotics in aquaculture. Probiotics. antimicrob. proteins. 10 (1), 98–109. doi: 10.1007/s12602-017-9312-8
Kavitha M., Raja M., Perumal P. (2018). Evaluation of probiotic potential of bacillus spp. isolated from the digestive tract of freshwater fish labeo calbasu (Hamilton, 1822). Aquacult. Rep. 11, 59–69. doi: 10.1016/j.aqrep.2018.07.001
Khan M. I. R., Choudhury T. G., Kamilya D., Monsang S. J., Parhi J. (2021). Characterization of bacillus spp. isolated from intestine of labeo rohita–towards identifying novel probiotics for aquaculture. Aquacult. Res. 52 (2), 822–830. doi: 10.1111/are.14937
Khan M. I. R., Kamilya D., Choudhury T. G., Tripathy P. S., Rathore G. (2021). Deciphering the probiotic potential of bacillus amyloliquefaciens COFCAU_P1 isolated from the intestine of labeo rohita through in vitro and genetic assessment. Probiotics. Antimicrob. Proteins, 1–13. doi: 10.1007/s12602-021-09788-2
Kuebutornye F. K. A., Abarike E. D., Lu Y. (2019). A review on the application of bacillus as probiotics in aquaculture. Fish. Shellfish. Immunol. 87, 820–828. doi: 10.1016/j.fsi.2019.02.010
Kuebutornye F. K., Abarike E. D., Lu Y., Hlordzi V., Sakyi M. E., Afriyie G., et al. (2020). Mechanisms and the role of probiotic bacillus in mitigating fish pathogens in aquaculture. Fish. Physiol. Biochem., 2020 (46), 819–841. doi: 10.1007/s10695-019-00754-y
Kuebutornye F. K., Lu Y., Abarike E. D., Wang Z., Li Y., Sakyi M. E. (2020). In vitro assessment of the probiotic characteristics of three bacillus species from the gut of Nile tilapia, oreochromis niloticus. Probiotics. antimicrob. proteins. 12 (2), 412–424. doi: 10.1007/s12602-019-09562-5
Lei X., Piao X., Ru Y., Zhang H., Péron A., Zhang H. (2015). Effect of bacillus amyloliquefaciens based direct-fed microbial on performance, nutrient utilization, intestinal morphology and cecal microflora in broiler chickens. Asian-Australasian J. Anim. Sci. 28 (2), 239. doi: 10.5713/ajas.14.0330
Li X., Ringø E., Hoseinifar S. H., Lauzon H. L., Birkbeck H., Yang D. (2019). The adherence and colonization of microorganisms in fish gastrointestinal tract. Rev. Aquaculture. 11 (3), 603–618. doi: 10.1111/raq.12248
Li M., Wang Y., Cui H., Li Y., Sun Y., Qiu H.-J. (2020). Characterization of lactic acid bacteria isolated from the gastrointestinal tract of a wild boar as potential probiotics. Front. vet. science. 7. doi: 10.3389/fvets.2020.00049
Makridis P., Kokou F., Bournakas C., Papandroulakis N., Sarropoulou E. (2021). Isolation of phaeobacter sp. from larvae of atlantic bonito (Sarda sarda) in a mesocosmos unit, and its use for the rearing of european seabass larvae (Dicentrarchus labrax l.). Microorganisms. 9 (1), 128. doi: 10.3390/microorganisms9010128
Mbozo A. B. V., Kobawila S. C., Anyogu A., Awamaria B., Louembe D., Sutherland J. P., et al. (2017). Investigation of the diversity and safety of the predominant bacillus pumilus sensu lato and other bacillus species involved in the alkaline fermentation of cassava leaves for the production of ntoba mbodi. Food control. 82, 154–162. doi: 10.1016/j.foodcont.2017.06.018
Medina M., Sotil G., Flores V., Fernández C., Sandoval N. (2020). In vitro assessment of some probiotic properties and inhibitory activity against yersinia ruckeri of bacteria isolated from rainbow trout oncorhynchus mykiss (Walbaum). Aquacult. Rep. 18, 100447. doi: 10.1016/j.aqrep.2020.100447
Meidong R., Doolgindachbaporn S., Jamjan W., Sakai K., Tashiro Y., Okugawa Y., et al. (2017). A novel probiotic bacillus siamensis B44v isolated from Thai pickled vegetables (Phak-dong) for potential use as a feed supplement in aquaculture. J. Gen. Appl. Microbiol. 63 (4), 246–253. doi: 10.2323/jgam.2016.12.002
Meidong R., Khotchanalekha K., Doolgindachbaporn S., Nagasawa T., Nakao M., Sakai K., et al. (2018). Evaluation of probiotic bacillus aerius B81e isolated from healthy hybrid catfish on growth, disease resistance and innate immunity of pla-mong pangasius bocourti. Fish. shellfish. Immunol. 73, 1–10. doi: 10.1016/j.fsi.2017.11.032
Midhun S. J., Neethu S., Vysakh A., Arun D., Radhakrishnan E., Jyothis M. (2017). Antibacterial activity and probiotic characterization of autochthonous paenibacillus polymyxa isolated from anabas testudineus (Bloch, 1792). Microb. pathog. 113, 403–411. doi: 10.1016/j.micpath.2017.11.019
Midhun S. J., Neethu S., Vysakh A., Radhakrishnan E., Jyothis M. (2018). Antagonism against fish pathogens by cellular Components/Preparations of bacillus coagulans (MTCC-9872) and it’s In vitro probiotic characterisation. Curr. Microbiol. 75 (9), 1174–1181. doi: 10.1007/s00284-018-1506-0
Midhun S. J., Neethu S., Vysakh A., Sunil M., Radhakrishnan E., Jyothis M. (2017). Antibacterial activity of autochthonous bacteria isolated from anabas testudineus (Bloch, 1792) and it’s in vitro probiotic characterization. Microb. pathog. 113, 312–320. doi: 10.1016/j.micpath.2017.10.058
Mukherjee A., Ghosh K. (2016). Antagonism against fish pathogens by cellular components and verification of probiotic properties in autochthonous bacteria isolated from the gut of an Indian major carp, catla catla (H amilton). Aquacult. Res. 47 (7), 2243–2255. doi: 10.1111/are.12676
Muñoz-Atienza E., Araújo C., Magadán S., Hernández P. E., Herranz C., Santos Y., et al. (2014). In vitro and in vivo evaluation of lactic acid bacteria of aquatic origin as probiotics for turbot (Scophthalmus maximus l.) farming. Fish. shellfish. Immunol. 41 (2), 570–580. doi: 10.1016/j.fsi.2014.10.007
Muñoz-Atienza E., Gómez-Sala B., Araújo C., Campanero C., Del Campo R., Hernández P. E., et al. (2013). Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 13 (1), 15. doi: 10.1186/1471-2180-13-15
Nayak S. (2010). Probiotics and immunity: a fish perspective. Fish. shellfish. Immunol. 29 (1), 2–14. doi: 10.1016/j.fsi.2010.02.017
Pinto A., Barbosa J., Albano H., Isidro J., Teixeira P. (2020). Screening of bacteriocinogenic lactic acid bacteria and their characterization as potential probiotics. Microorganisms 8 (3), 393. doi: 10.3390/microorganisms8030393
Prieto M., O’Sullivan L., Tan S., McLoughlin P., Hughes H., Gutierrez M., et al. (2014). In vitro assessment of marine bacillus for use as livestock probiotics. Mar. Drugs 12 (5), 2422–2445. doi: 10.3390/md12052422
Ramesh D., Vinothkanna A., Rai A. K., Vignesh V. S. (2015). Isolation of potential probiotic bacillus spp. and assessment of their subcellular components to induce immune responses in labeo rohita against aeromonas hydrophila. Fish. shellfish. Immunol. 45 (2), 268–276. doi: 10.1016/j.fsi.2015.04.018
Romero J., Ringø E., Merrifield D. L. (2014). “The gut microbiota of fish,” in Aquaculture nutrition: Gut health, probiotics and prebiotics Oxford, (Wiley-Blackwell Publishing), 101, 75–100. doi: 10.1002/9781118897263.ch4
Rungsirivanich P., Supandee W., Futui W., Chumsai-Na-Ayudhya V., Yodsombat C., Thongwai N. (2020). Culturable bacterial community on leaves of Assam tea (Camellia sinensis var. assamica) in Thailand and human probiotic potential of isolated bacillus spp. Microorganisms 8 (10), 1585. doi: 10.3390/microorganisms8101585
Sahoo T. K., Jena P. K., Nagar N., Patel A. K., Seshadri S. (2015). In vitro evaluation of probiotic properties of lactic acid bacteria from the gut of labeo rohita and catla catla. Probiotics. antimicrob. proteins. 7 (2), 126–136. doi: 10.1007/s12602-015-9184-8
Samson J. S., Choresca C. H., Quiazon K. M. A. (2020). Selection and screening of bacteria from African night crawler, eudrilus eugeniae (Kinberg, 1867) as potential probiotics in aquaculture. World J. Microbiol. Biotechnol. 36 (1), 16. doi: 10.1007/s11274-019-2793-8
Santos R. A., Oliva-Teles A., Pousão-Ferreira P., Jerusik R., Saavedra M. J., Enes P., et al. (2021). Isolation and characterization of fish-gut bacillus spp. as source of natural antimicrobial compounds to fight aquaculture bacterial diseases. Mar. Biotechnol. 23 (2), 276–293. doi: 10.1007/s10126-021-10022-x
Sharifuzzaman S., Rahman H., Austin D. A., Austin B. (2018). Properties of probiotics kocuria SM1 and rhodococcus SM2 isolated from fish guts. Probiotics. antimicrob. proteins. 10 (3), 534–542. doi: 10.1007/s12602-017-9290-x
Shinde T., Vemuri R., Shastri M. D., Perera A. P., Tristram S., Stanley R., et al. (2019). Probiotic bacillus coagulans MTCC5856 spores exhibit excellent in vitro functional efficacy in simulated gastric survival, mucosal adhesion and immunomodulation. J. Funct. foods. 52, 100–108. doi: 10.1016/j.jff.2018.10.031
Verschuere L., Rombaut G., Sorgeloos P., Verstraete W. (2000). Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. Rev. 64 (4), 655–671. doi: 10.1128/MMBR.64.4.655-671.2000
Yamashita M. M., Ferrarezi J. V., GdV P., Bandeira G., Côrrea da Silva B., SA P., et al. (2020). Autochthonous vs allochthonous probiotic strains to rhamdia quelen. MicrobialPathogenesis 139, 103897. doi: 10.1016/j.micpath.2019.103897
Yılmaz S., Yılmaz E., Dawood M. A., Ringø E., Ahmadifar E., Abdel-Latif H. M. (2021). Probiotics, prebiotics, and synbiotics used to control vibriosis in fish: A review. Aquaculture 737514, 73–75. doi: 10.1016/j.aquaculture.2021.737514
Zhou S., Song D., Zhou X., Mao X., Zhou X., Wang S., et al. (2019). Characterization of bacillus subtilis from gastrointestinal tract of hybrid hulong grouper (Epinephelus fuscoguttatus× e. lanceolatus) and its effects as probiotic additives. Fish. shellfish. Immunol. 84, 1115–1124. doi: 10.1016/j.fsi.2018.10.058
Keywords: Vibriosis, probiotic, Bacillus spp., safety assessment, adhesion
Citation: Jlidi M, Akremi I, Ibrahim AH, Brabra W, Ali MB and Ali MB (2022) Probiotic properties of Bacillus strains isolated from the gastrointestinal tract against pathogenic Vibriosis. Front. Mar. Sci. 9:884244. doi: 10.3389/fmars.2022.884244
Received: 06 March 2022; Accepted: 27 June 2022;
Published: 15 August 2022.
Edited by:
Xiangli Tian, Ocean University of China, ChinaReviewed by:
Vikash Kumar, Central Inland Fisheries Research Institute (ICAR), IndiaThavasimuthu Citarasu, Manonmaniam Sundaranar University, India
Copyright © 2022 Jlidi, Akremi, Ibrahim, Brabra, Ali and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mouna Jlidi, jlidimanno@yahoo.fr; Mamdouh Ben Ali, mamdouh.benali@cbs.rnrt.tn
 Mouna Jlidi
Mouna Jlidi Ismahen Akremi1,2
Ismahen Akremi1,2  Adel Haj Ibrahim
Adel Haj Ibrahim