Microbial community characteristics of the intestine and gills of medium-form populations of Sthenoteuthis oualaniensis in the South China Sea
- 1South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2Key Laboratory of South China Sea Fishery Resources Exploitation & Utilization, Ministry of Agriculture and Rural Affairs, Guangzhou, China
- 3Guangdong Provincial Key Laboratory of Fishery Ecology and Environment, Guangzhou, China
Sthenoteuthis oualaniensis (purpleback squid) is an excellent biological resource in the South China Sea. However, the microbiological characteristics of this South China Sea squid, especially those of the medium-form of different sexes and gonadal maturities, are poorly understood. In this study, the characteristics of the bacterial community in the intestinal and gill tissues of female and male S. oualaniensis with different gonadal maturities, collected from the Nansha Sea of China in spring 2020, were analyzed. The results showed that Tenericutes, Proteobacteria, Firmicutes, and Bacteroidetes were the dominant phyla in the intestinal microbial samples of female immature gonad (FN), male immature gonad (MN), and male sexual maturity (MY) samples of the S. oualaniensis populations. Proteobacteria, Firmicutes, Bacteroidetes, and Tenericutes were the dominant phyla in the intestinal microbial sample of the female sexual maturity (FY) group. The microbial community in the gills differed from that of the intestinal flora. The dominant phyla in the gill samples were Proteobacteria, Firmicutes, and Bacteroidetes, regardless of sex or gonadal maturity. According to the random forest analysis, the gill samples had significantly (p<0.001) more Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in the top 20 than the intestinal samples. Correlation analysis showed that the mantle length of S. oualaniensis was significantly (p<0.05) negatively correlated with the abundance of Tenericutes, but significantly (p<0.01) positively correlated with Firmicutes and Bacteroidetes. The mantle length of FY was significantly (p<0.05) longer than that of the other types. The results indicated that the differences in the intestinal bacterial community were related to the growth and feeding characteristics of S. oualaniensis of different sexes and maturities.
Introduction
Sthenoteuthis oualaniensis (Lesson, 1830) is a warm-water oceanic cephalopod belonging to the class Cephalopoda, order Oegopsida, and family Ommastrephidae (Dong, 1988). It is mainly distributed in the subtropical and equatorial waters of the Indian and Pacific Oceans. It is most abundant in the South China Sea and northwestern Indian Ocean (Zhang P et al., 2015). Squid from the South China Sea, commonly known as deep-sea red squid, are one of the most promising biological resources in the South China Sea because of their abundance and short life cycle (Zhang et al., 2010). Sthenoteuthis oualaniensis individuals were divided into three categories, namely medium-form population, dwarf-form population, and X3-form population, based on characteristics such as mantle length, dorsal photophore, and statolith morphology (Jiang et al., 2015). Among them, medium-form populations are one of the main squid groups from the South China Sea (Zhang P et al., 2015). At present, research on S. oualaniensis has mainly focused on resource assessment (Chen et al., 2013; Zhang J et al., 2015), growth and feeding (Gong et al., 2018), morphological variations and discrimination (Zhu et al., 2019), genetic diversity (Li M et al., 2019), and nutritional processing (Huang et al., 2020). There have been few reports on the symbiotic relationships of this squid with microorganisms.
Intestinal microbes are an essential part of the digestive system of invertebrates. Research on the composition and determining factors of invertebrate intestinal microbiota, such as that of cephalopods, is essential for understanding their symbiotic mechanism (Kang et al., 2022). In general, invertebrate microbial communities are relatively simple (Webster et al., 2010). Although invertebrates are often exposed to a large number of microorganisms in their habitats, only a few species of bacteria are found in their digestive tracts (Kang et al., 2022). The gill is an essential organ for gaseous exchange. The microbial composition of invertebrate intestines and gills varies with the changes in several factors, such as feeding habits, bait, and various environmental factors (Jin et al., 2017; Duan et al., 2020). However, little is known about the microbiomes of invertebrates, particularly that of S. oualaniensis. Environmental microbes, water salinity, water temperature, bait, drugs, different physiological conditions, and different developmental phases affect the microbial community, especially the microbial species diversity (Wang et al., 2018; Li Y M et al., 2019; Sun et al., 2022).
The microbiological characteristics of the South China Sea squid, particularly those of the medium form of different sexes and gonadal maturities, are poorly understood. Therefore, we aimed to analyze the characteristics of the bacterial community in the intestinal and gill tissues of females and males with different gonadal maturities in medium-form populations of S. oualaniensis collected from the Nansha Sea of China in the spring of 2020. This study aimed to reveal the (a) differences in the intestinal and gill microbial communities in the medium-form squid populations of S. oualaniensis and the (b) relationship among the microbial communities of the different sexes of this species at different gonadal maturities. These results will provide information on the biological characteristics of the microbial communities of S. oualaniensis in the South China Sea.
Materials and methods
Sample collection and processing
South China Sea squids were caught using a light cover net at stations N1 (11.00°N, 114.00°E) and N2 (9.00°N, 114.00°E) in the Nansha area of China, between May 21 and 27, 2020.
Male and female purpleback squid are distinguished by whether or not their left fourth wrist is stemmed and the difference in the gonadal structure (Wang and Chen, 2005; Zhang P et al., 2015). Immature female gonad (FN), female sexual maturity (FY), immature male gonad (MN), and male sexual maturity (MY) samples of medium-form populations of squid were randomly collected. Six squid samples from each group were selected from sites N1 and N2.
The mantle length of S. oualaniensis was measured using a measuring plate with an accuracy of 1 mm, and the body mass was weighed using a weighing scale. The surface of S. oualaniensis was sterilized using 75% alcohol and the intestine and gill samples of different sexes with different gonadal maturities were collected. Each group included six replicates of intestinal and gill samples. The intestinal and gill samples were stored in sterile cryotubes with preservation solution, consisting of 75% absolute alcohol and 1% 0.5 M EDTA, at -20 °C for future analysis.
Ethics statement
The protocols for collecting and handling the animals in this study were approved by the Animal Care and Use Committee of the South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (SCSFRI-CAFS, No. nhdf2022-03). All experimental animal protocols were implemented in accordance with national and institutional guidelines for the care and use of laboratory animals at the SCSFRI-CAFS.
DNA extraction and PCR amplification
Before microbial molecular analysis, intestinal and gill samples of FN, FY, MN, and MY were removed from the preservation solution. The bacterial DNA was extracted using a MoBio PowerFecal DNA Isolation Kit (MoBio, Carlsbad, CA). The primers 515F (GTGCCAGCMGCCGC GG) and 806R (GGACTACHVGGGTWTCTAAT) were used to amplify the V4 region of the 16S ribosomal RNA (Duan et al., 2020). The PCR mixture comprised of a 20 μL reaction volume containing 10 ng of template DNA, 0.4 μL of FastPfu Polymerase, 0.8 μL of each primer (5 μM), 2 μL of 2.5 mM dNTPs, 4 μL of 5× FastPfu Buffer, and distilled water. The PCR conditions were as follows: 95 °C for 2 min; 25 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 5 min. Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA)
Library construction and sequencing
Purified PCR products were quantified using Qubit® 3.0 (Invitrogen, Waltham, MA), and the 24 amplicons with differing barcodes were mixed equally. According to Illumina genomic DNA library preparation procedure, the pooled DNA products were used to construct the Illumina paired-end library. Paired-end sequencing was performed on an Illumina HiSeq 2500 platform (Mingke Biotechnology Co., Ltd., Hangzhou, China).
16S rRNA sequence analysis
UPARSE was used to cluster operational taxonomic units (OTUs) with 97% similarity (Amato et al., 2013). Rarefaction analysis was conducted using Mothur software to calculate the diversity indices, including the Chao1, ACE, Simpson, and Shannon diversity indices (Schloss et al., 2011). A Venn diagram was drawn to reveal the unique and shared OTUs in the different samples (Fouts et al., 2012). Then, using the Ape package, the beta diversity was analyzed using principal coordinate analysis (PCoA; Paradis et al., 2004). The microbial community composition in the intestinal and gill tissues of the squid samples was determined at the phylum and genus levels. Correlation and differential networks were constructed using Cytoscape software according to the relative abundance of individual OTUs (http://www.cytoscape.org/). The linear discriminant analysis (LDA) effect size (LEfSe) was used for quantitative analysis (Segata et al., 2011). Random forest analyses were performed using the R random forest and ggplot 2 packages at Kyoto Encyclopedia of Genes and Genomes (KEGG) level 3.
Accession number
The raw data in this study was deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database. The accession number was PRJNA822533.
Statistical analysis
The correlation between mantle length of S. oualaniensis and relative abundance of the dominant bacteria was analyzed using Pearson correlation analysis in SPSS software. Statistical significance was set at p<0.05.
Results
Mantle length and body mass of Sthenoteuthis oualaniensis
The mantle length and body mass of FY were significantly (p<0.05) higher than that of the other types, with an average mantle length and body mass of 190.8 mm and 342.3 g, respectively (Figures 1, 2). The MY group’s average mantle length and body mass were 143.7 mm and 139.8 g, respectively. The mantle length and body length of MY were significantly (p<0.05) higher than that of FN and MN. The average mantle length and body mass of FN were 119.5 mm and 69.7 g, respectively, and those of MN were 112.5 mm and 58.5 g, respectively. There were no significant (p>0.05) differences in mantle length or body mass between the immature gonad groups.
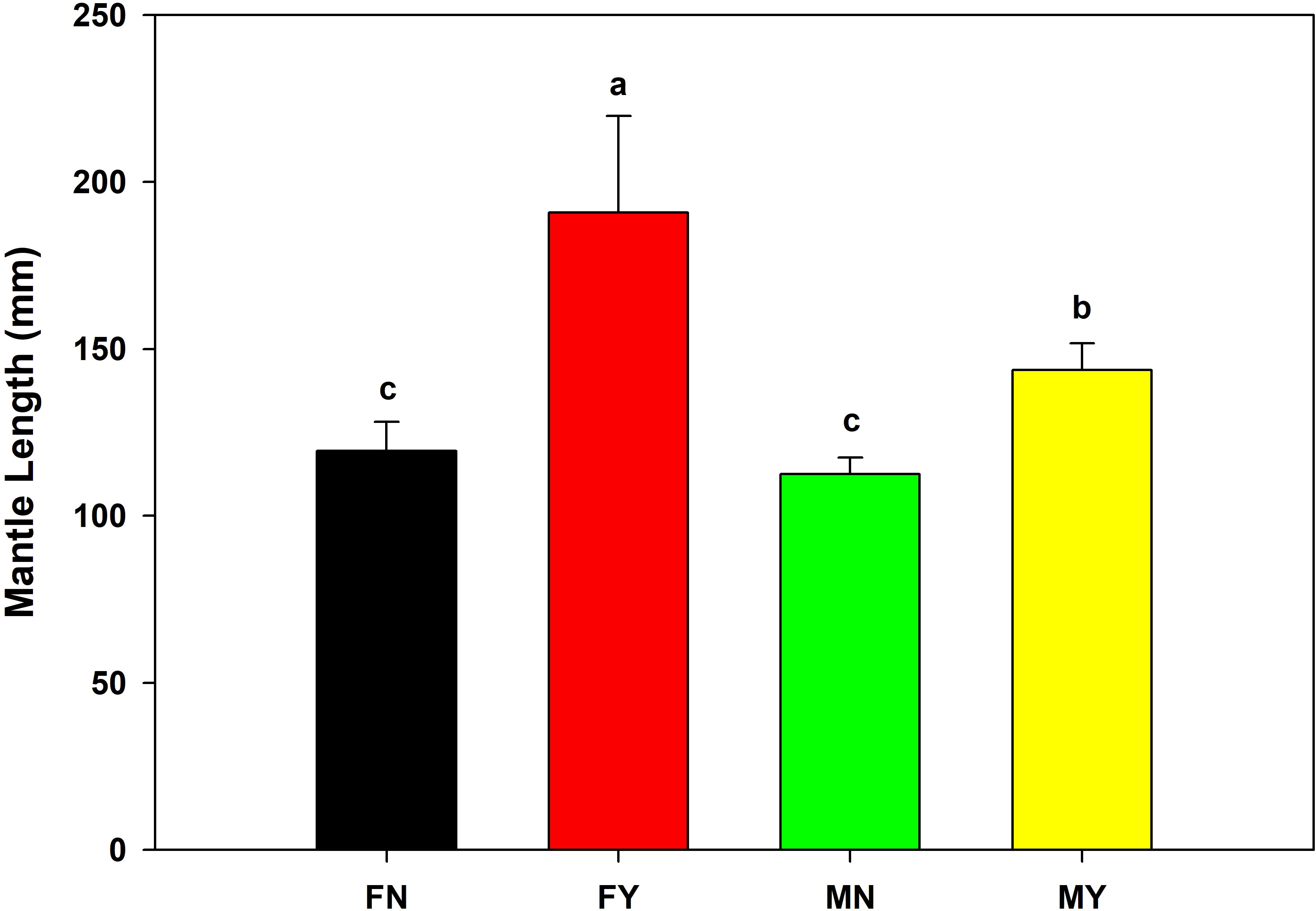
Figure 1 Mantle lengths of medium-form populations of Sthenoteuthis oualaniensis. Significant differences are indicated by different letters (p < 0.05).
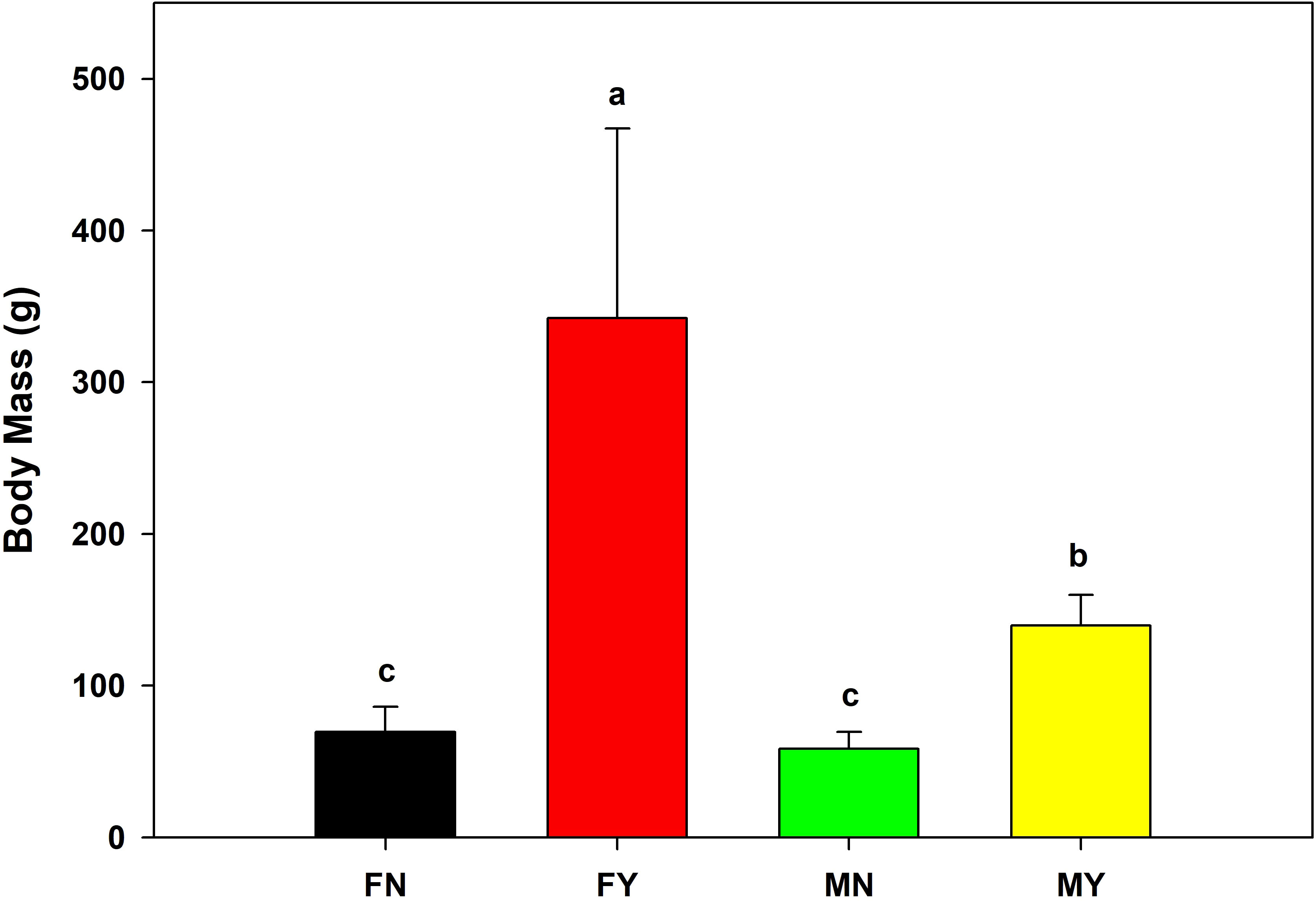
Figure 2 Body masses of medium-form populations of Sthenoteuthis oualaniensis. Significant differences are indicated by different letters (p < 0.05).
Microbial richness and diversity
Alpha diversity indices, including the Chao1, ACE, Simpson, and Shannon diversity indices, were calculated (Figure 3). The Chao1 index of the gill samples of immature female gonad (FNG) was significantly higher than that of the male sexual maturity (MYG). The Simpson indices of the intestinal samples of female sexual maturity (FYI) were significantly lower than that of the other samples, except for that of the intestinal samples of immature female gonad (FNI). The Shannon index of the FYI group was the highest.
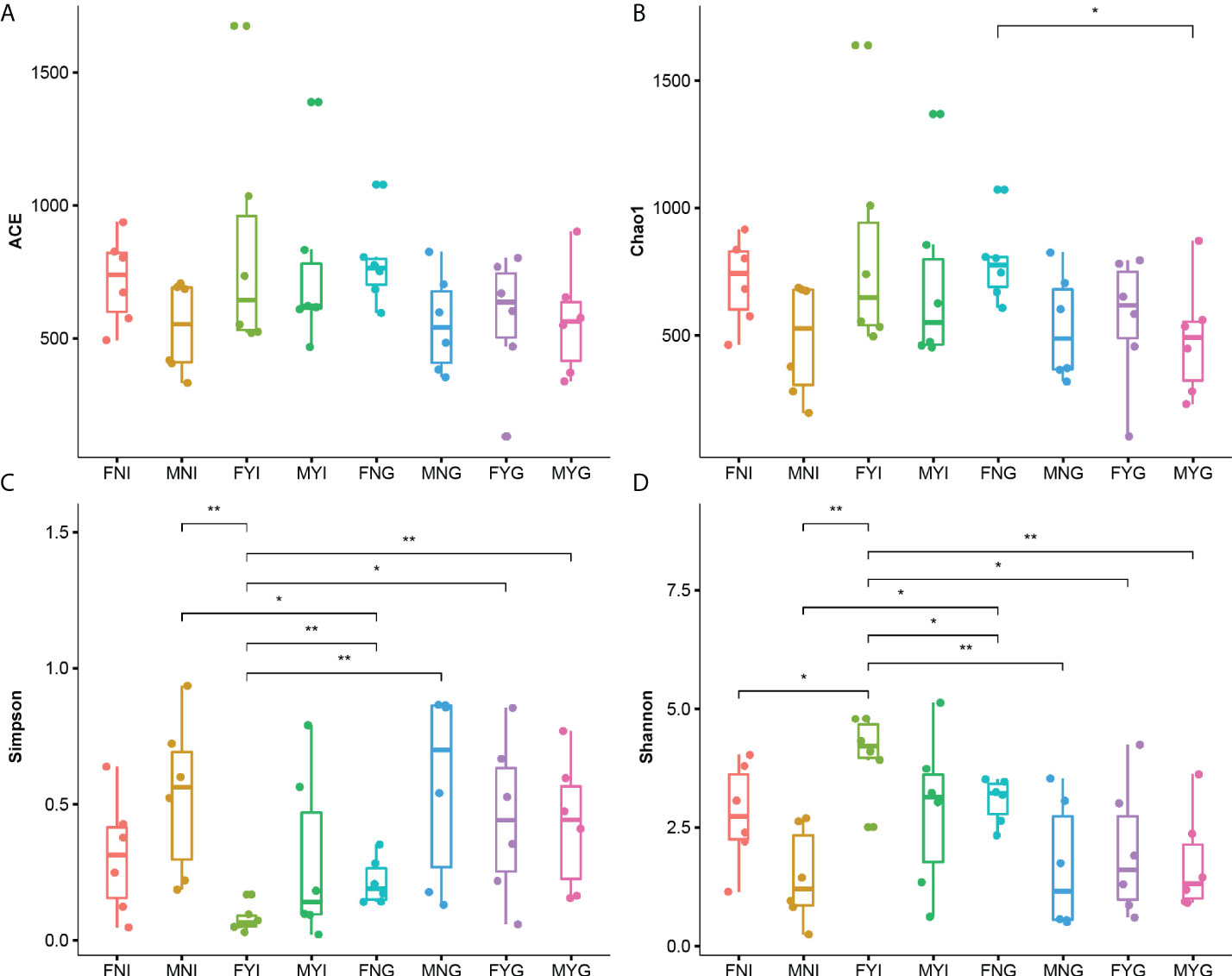
Figure 3 Microbial diversity indices in the intestine and gill microbial samples of medium-form populations of Sthenoteuthis oualaniensis. (A) Observed, (B) Chao1 indices, (C) Simpson indices, and (D) Shannon indices. FNI represents the intestine samples of female immature gonad, MNI represents the intestine samples of male immature gonad, FYI represents the intestine samples of female sexual maturity, and MYI represents the intestine samples of male sexual maturity, FNG represents the gill samples of female immature gonad, MNG represents the gill samples of male immature gonad, FYG represents the gill samples of female sexual maturity, and MYG represents the gill samples of male sexual maturity. *Indicates the significant difference between two samples (p < 0.05) and **indicates the extremely significant difference between two samples (p < 0.01).
A total of 676 OTUs were shared among the four intestinal groups as indicated by the Venn diagram (Figure 4A). Among them, the FYI group had the highest number (478 OTUs) of unique OTUs, while those of the intestinal samples of male sexual maturity (MYI; 331 OTUs), FNI (159 OTUs), and the intestinal samples of immature male gonad (MNI; 44 OTUs) were lower. Figure 4B shows that 592 OTUs were shared among the four gill groups. The FNG group had the highest number (342 OTUs) of unique OTUs, while those of the gill samples of female sexual maturity (FYG; 182 OTUs), MYG (173 OTUs), and the gill samples of immature male gonad (MNG; 97 OTUs) were lower. The intestinal and gill bacterial groups of S. oualaniensis were clustered together, according to the PCoA-based weighted UniFrac distances (Figure 5). Most of the intestinal bacterial groups were located in the negative axis region of the abscissa, and most of the gill bacterial groups were located in the positive axis region.
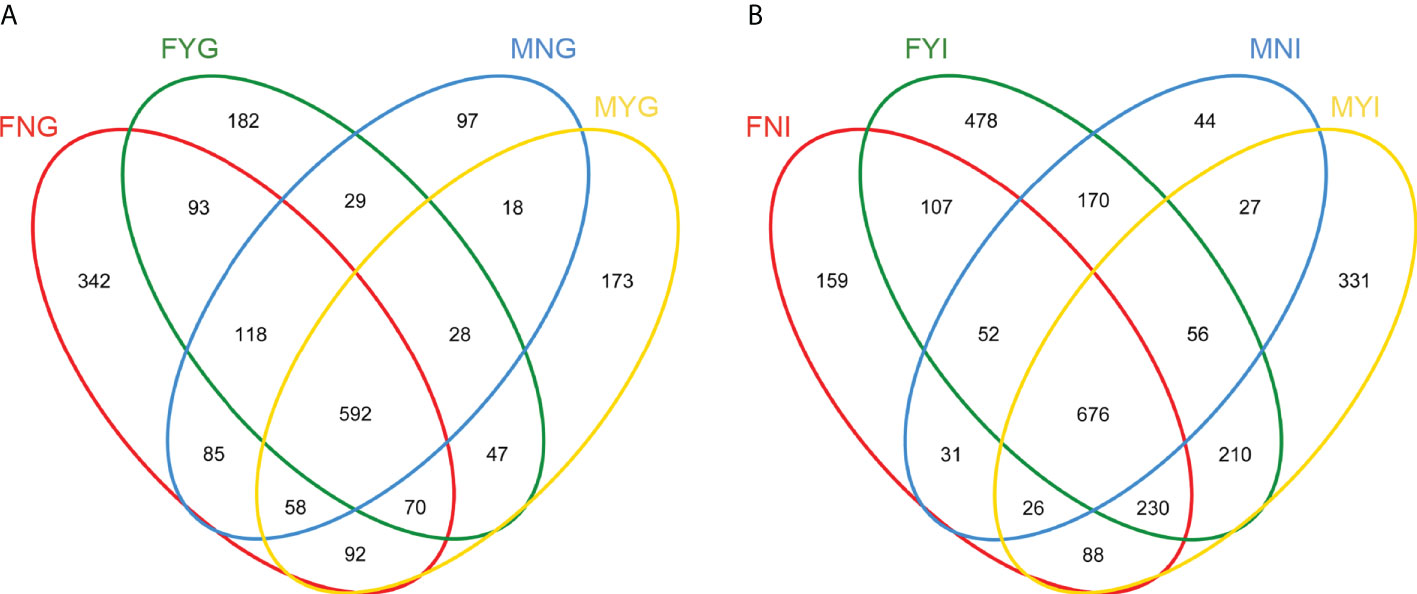
Figure 4 Venn diagram of the intestine (A) and gill (B) microbial samples of medium-form populations of Sthenoteuthis oualaniensis..
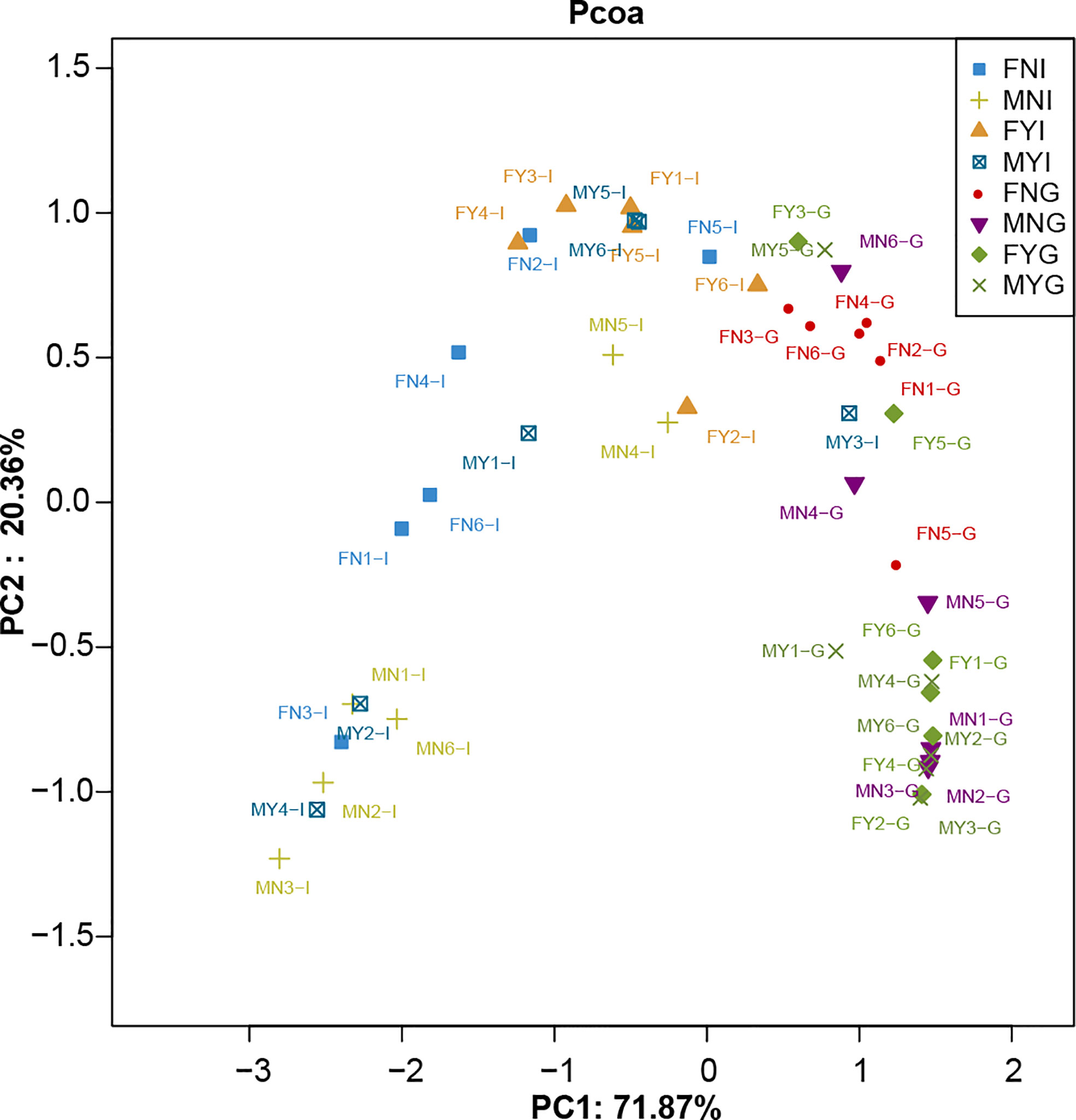
Figure 5 Principal coordinate analysis (PCoA) of the intestine and gill microbial samples of medium-form populations of Sthenoteuthis oualaniensis based on weighted UniFrac metrics.
Microbial community
A total of 40 phyla and 963 genera were identified in the intestinal and gill microbial samples from the S. oualaniensis populations. Tenericutes, Proteobacteria, Firmicutes, and Bacteroidetes were the dominant phyla in the FNI, MNI, and MYI groups, whereas Proteobacteria, Firmicutes, and Bacteroidetes were the dominant phyla in the FYG, MNG, and MYG groups (Figure 6).
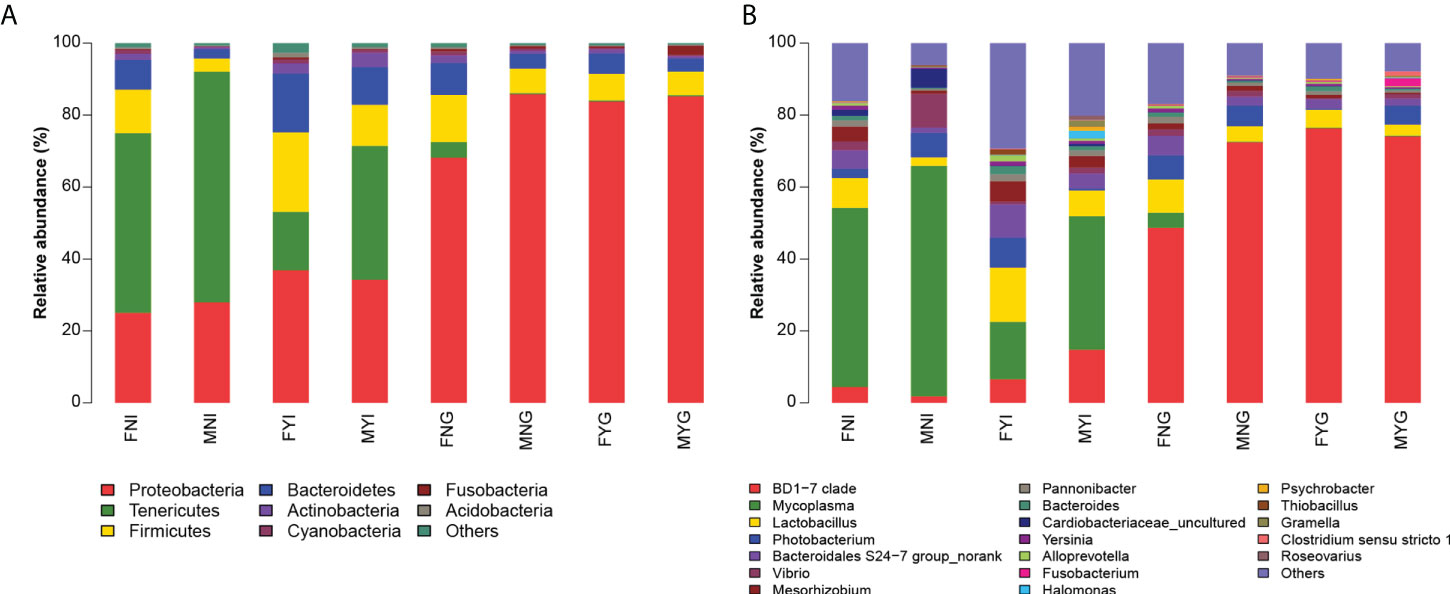
Figure 6 Bacterial community composition in intestinal and gill tissues of medium-form squid. (A) Phylum level, (B) Genus level.
In the FNI group, Tenericutes (relative abundance of 49.60% ± 25.12%), Proteobacteria (24.87% ± 15.20%), Firmicutes (12.04% ± 6.88%), and Bacteroidetes (8.19% ± 4.43%) were the dominant phyla. At the genus level, the dominant bacteria were Mycoplasma (relative abundance of 49.45% ± 25.16%), Lactobacillus (8.33% ± 4.87%), and Bacteroidales S24-7 group (5.10% ± 2.77%). In the MNI group, Tenericutes (relative abundance of 63.95% ± 28.54%), Proteobacteria (27.89% ± 21.01%), Firmicutes (3.71% ± 4.59%), and Bacteroidetes (2.58% ± 2.65%) were the dominant phyla. The dominant genera in the MNI group were Mycoplasma (relative abundance of 63.89% ± 28.60%), Vibrio (9.35% ± 13.28%), Photobacterium (6.80% ± 12.55%), Cardiobacteriaceae (5.22% ± 2.41%), Lactobacillus (2.41% ± 2.97%), and Bacteroidales S24-7 group (1.47% ± 1.76%). In the FYI group, Proteobacteria (relative abundance of 36.62% ± 18.00%), Firmicutes (21.94% ± 11.55%), Bacteroidetes (16.25% ± 5.69%), and Tenericutes (16.16% ± 11.58%) were the dominant phyla. At the genus level, the dominant bacteria were Mycoplasma (relative abundance of 15.83% ± 11.49%), Lactobacillus (15.00% ± 8.26%), Bacteroidales S24-7 group (9.21% ± 4.49%), Photobacterium (8.20% ± 16.91%), and BD1-7 clade (6.56% ± 7.26%). In the MYI group, Tenericutes (relative abundance of 36.92% ± 37.06%), Proteobacteria (33.94% ± 22.33%), Firmicutes (11.38% ± 11.29%), and Bacteroidetes (10.30% ± 8.47%) were the dominant phyla. The dominant genera were Mycoplasma (relative abundance of 36.78% ± 37.18%), BD1-7 clade (14.67% ± 20.41%), Lactobacillus (7.14% ± 8.59%), Bacteroidales S24-7 group (4.11% ± 5.24%), and Mesorhizobium (3.19% ± 3.46%).
Proteobacteria (relative abundance of 67.75% ± 8.38%), Firmicutes (13.04% ± 3.43%), Bacteroidetes (8.78% ± 2.63%), and Tenericutes (4.34% ± 6.16%) were the main bacteria in the FNG group. The dominant genera were BD1-7 clade (relative abundance of 48.41% ± 7.15%), Lactobacillus (9.21% ± 2.55%), Photobacterium (6.62% ± 9.29%), and Bacteroidales S24-7 group (5.38% ± 1.74%). In the MNG groups, Proteobacteria (relative abundance of 83.56% ± 14.57%), Firmicutes (6.84% ± 6.92%), and Bacteroidetes (4.01% ± 4.13%) were the dominant bacteria. At the genus level, the dominant bacteria were BD1-7 clade (relative abundance of 72.22% ± 29.74%), Photobacterium (5.71% ± 13.20%), Lactobacillus (4.25% ± 4.50%), and Bacteroidales S24-7 group (2.50% ± 2.56%). The bacterial population of the FYG group was composed of Proteobacteria (relative abundance of 83.52% ± 17.42%), Firmicutes (7.46% ± 8.36%), and Bacteroidetes (5.55% ± 6.02%) as the dominant bacteria. The dominant genera were BD1-7 clade (relative abundance of 76.12% ± 24.46%), Lactobacillus (5.01% ± 5.59%), and Bacteroidales S24-7 group (2.64% ± 2.65%). The dominant bacterial phyla in the MYG group were Proteobacteria (relative abundance of 85.10% ± 16.69%), Firmicutes (6.55% ± 7.57%), and Bacteroidetes (3.54% ± 4.91%). At the genus level, the dominant bacteria were BD1-7 clade (relative abundance of 73.97% ± 30.04%), Photobacterium (5.20% ± 12.58%), Lactobacillus (3.09% ± 5.21%), and Bacteroidales S24-7 group (1.96% ± 2.96%).
Differential analysis
Differential abundance analysis of the bacterial taxa in the intestines and gills of medium-form squid was performed using LEfSe. Only the differences in the seven groups are shown in Figure 7A. In the intestinal bacterial community, the abundances of 0, 7, 26, and 9 bacterial groups were enriched in the FNI, MNI, FYI, and MYI groups, respectively, while 2, 1, 3, and 2 bacterial groups were enriched in the FNG, MNG, FYG, and MYG groups, respectively.
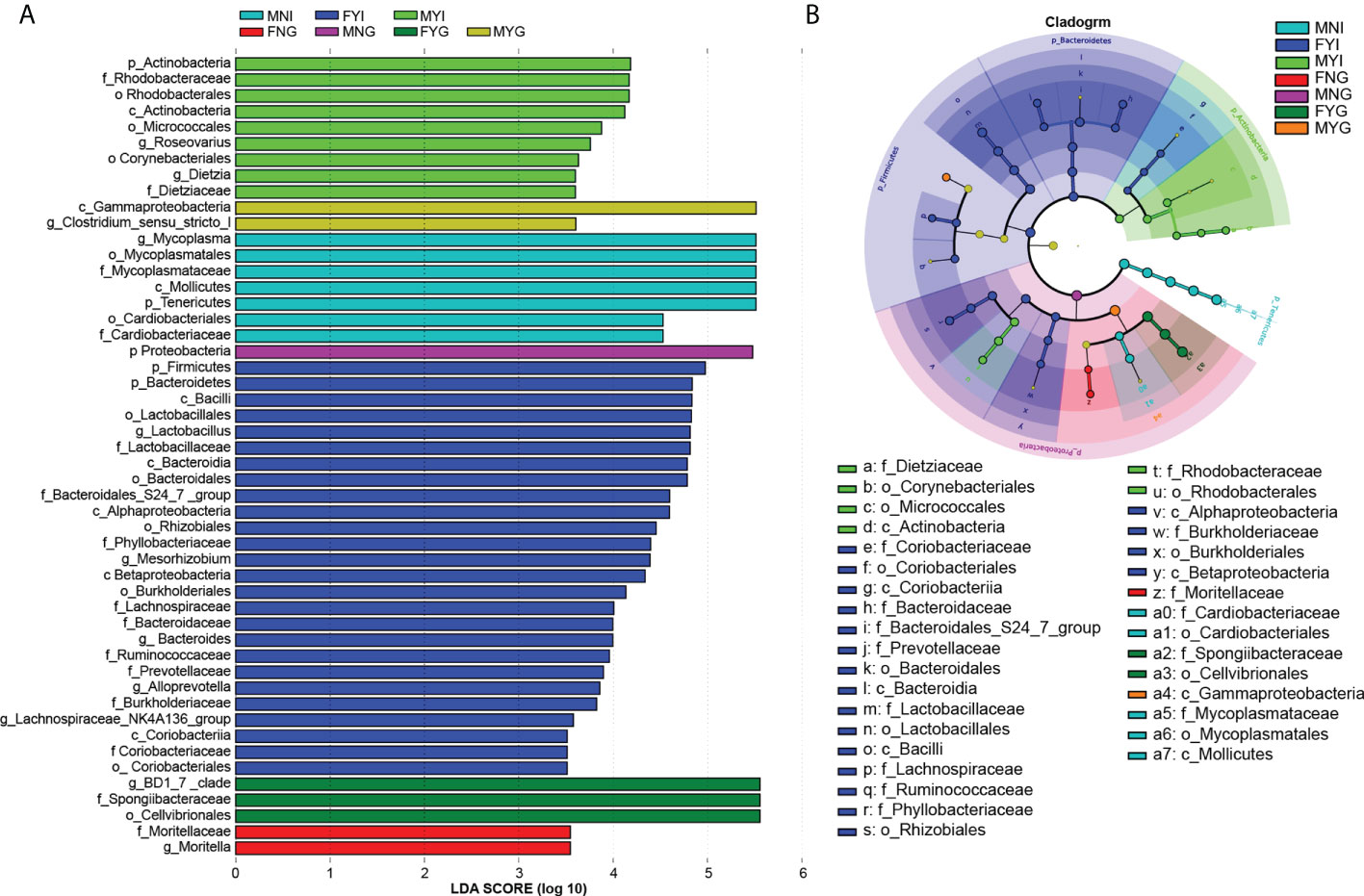
Figure 7 Inter-group variation of intestinal and gill tissues of medium-form squid. (A) LDA score; (B) LEfSe cladogram.
In detail, one class, two orders, and two families were enriched in the MNI group, including Mycoplasmataceae (from the class to the family level) and Cardiobacteriaceae (from the order to the family level). In the MYI group, one class, three orders, and two families, including Dietziaceae (from the class to the family level), Micrococcales (from the class to the order level), and Rhodobacteraceae (from the order to the family level), were enriched. Three classes, four orders, and eight families were enriched in the FYI group, namely Coriobacteriaceae (from the class to the family level), Bacteroidaceae (from the class to the family level), Bacteroidales S24-7 group (from the class to the family level), Prevotellaceae (from the class to the family level), Lactobacillaceae (from the class to the family level), Lachnospiraceae (from the family level), Ruminococcaceae (from the family level), and Phyllobacteriaceae (from the order to the family level). Moritellaceae was enriched in the FNG group. Gammaproteobacteria was also enriched in the MYG group. One class and one family were enriched in the FYG group, including Spongiibacteraceae (Figure 7B).
Network analyses
Comparing the microbiological characteristics among the medium forms of different sexes with different gonadal maturities showed several variations in the relative abundances of phyla. The dominant phyla were Proteobacteria and Tenericutes. Except for OTU1 (belonging to Proteobacteria) and OTU2 (belonging to Tenericutes), the other bacteria showed positive interactions with each other (Figure 8). Based on differential network analyses, the genus Mycoplasma and the family Cardiobacteriaceae were dominant in the MNI group. Class Bacilli, the families Bacteroidales S24-7 group and Lachnospirceae, and genera Lactobacillus, Bacteroides, Mesorhizobium, and Alloprevotella were dominant in FYI. The family Rhodobacteraceae and the class Actinobacteria were dominant in MYI. The family Moritellaceae was more prevalent in the FNG when comparing the gill microbiota. Only Proteobacteria was dominant in MNG. The genus BD1-7 clade was dominant in FYG. The class Gammaproteobacteria was dominant in the MYG (Figure 9).
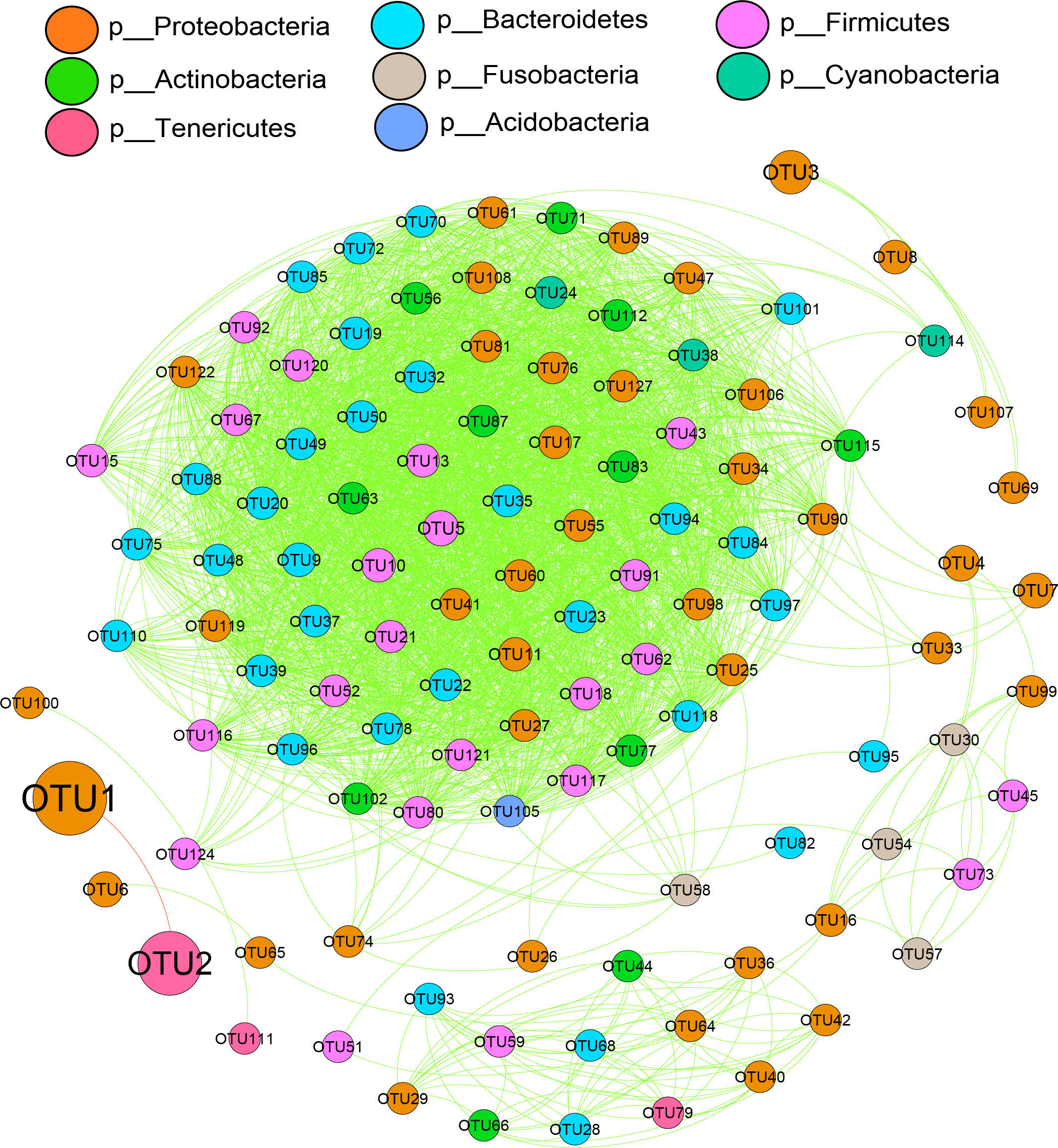
Figure 8 Correlation network analyses of the microbial communities of medium-form squid at the bacterial phyla level. The node size represents the abundance of each phylum. The green lines between phyla indicate a positive relationship, while the red lines indicate a negative relationship.
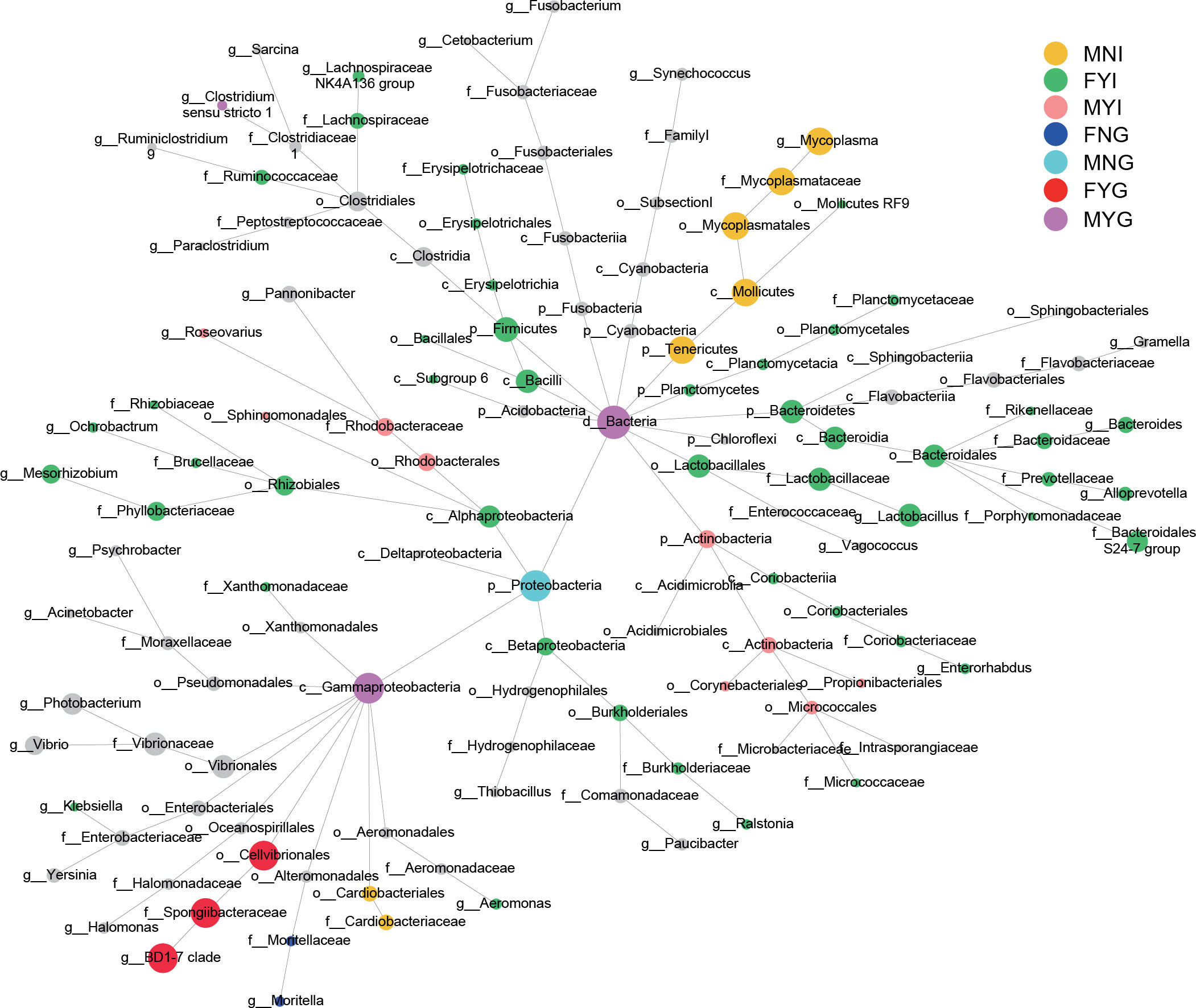
Figure 9 Differential network analyses of the intestine and gill bacterial communities of medium-form squid. The node indicates the species classification, and its size shows the abundance. Among them, the remaining colors show significant differences in the corresponding groups, and the black nodes represent no significant difference.
Functional prediction
The predicted functions of the intestinal and gill bacterial communities of the squid of different sexes and gonadal maturities were analyzed using the PICRUSt package. The gill samples had a significantly (p<0.001) higher level of the top 20 KEGG pathways than the intestinal samples, including ‘alpha-linoleic acid metabolism’, ‘stilbenoid, diarylheptanoid, and gingerol biosynthesis’, ‘xylene degradation’, ‘basal transcription’, ‘PPAR signaling pathway’, ‘geraniol degradation’, ‘arachidonic acid metabolism’, ‘steroid hormone biosynthesis’, ‘bile secretion’, ‘bisphenol degradation’, ‘fatty acid metabolism’, ‘Vibrio cholerae pathogenic cycle’, ‘dioxin degradation’, ‘lysine degradation’, ‘beta-alanine metabolism’, ‘synthesis and degradation of ketone bodies’, ‘adipocytokine signaling pathway’, ‘caprolactam degradation’, ‘benzoate degradation’, and ‘limonene and pinene degradation’ (Figure 10). The pathways among the top 20 were mainly related to the degradation of environmental organic matter and fatty acid metabolism. There were no significant differences between the gill samples. MNI had a lower level of most pathways, especially ‘bile secretion’.
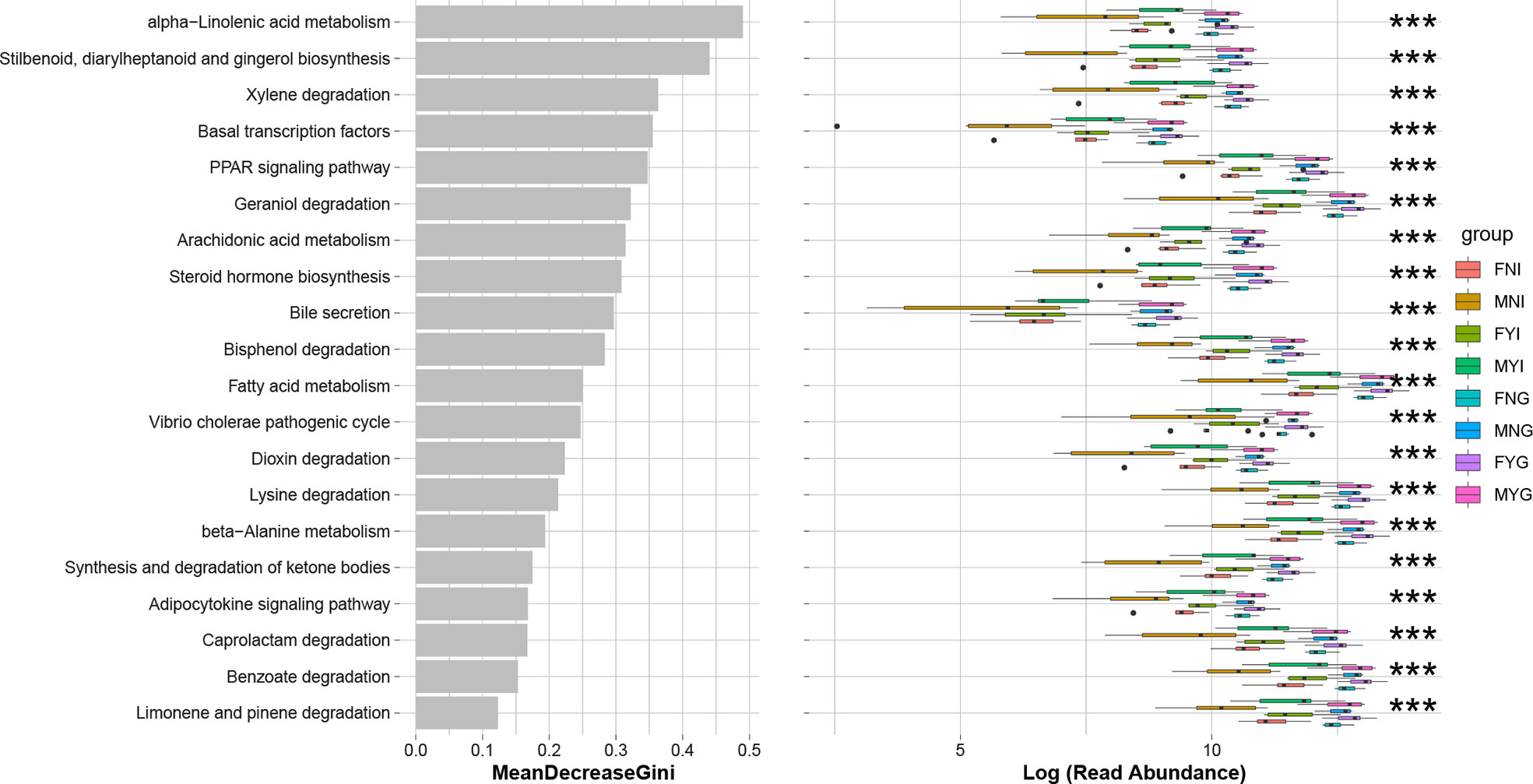
Figure 10 Prediction functions according to KEGG pathways in the bacterial communities of medium-form squid. *** indicates the significant difference between samples (p < 0.001).
Correlation analysis
Correlation analysis showed that the mantle length of S. oualaniensis was significantly (p<0.05) negatively correlated with the relative abundance of Tenericutes, but significantly (p<0.01) positively correlated with those of Firmicutes and Bacteroidetes (Table 1). At the genus level, mantle length had a significant (p<0.05) negative correlation with the relative abundance of Mycoplasma; however, it was significantly (p<0.05) positively correlated with Lactobacillus and had a highly significant (p<0.01) positive relationship with Bacteroidales S24-7 group (Table 2).
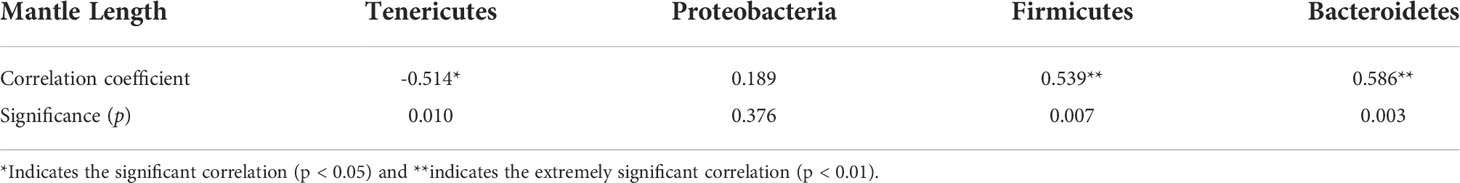
Table 1 Correlation analysis between the mantle length of Sthenoteuthis oualaniensis and the relative abundance of dominant intestinal bacteria at the phylum level.

Table 2 Correlation analysis between the mantle length of Sthenoteuthis oualaniensis and the relative abundance of dominant intestinal bacteria at the genus level.
Discussion
Microbial community characteristics in intestine and gill
Intestinal microbes are an important part of the digestive system, and gills are important organs for gaseous exchange. Few studies have focused on the cephalopod intestinal and gill microbial community characteristics. The intestinal microflora of Octopus mimus were investigated using a 16S rDNA clone library (Iehata et al., 2015). Strugnell and Nishiguchi (2007) analyzed the cephalopod intestinal microbial community in free-living and captive Octopus minor paralarvae using next-generation sequencing. A recent study confirmed the microbial composition of the digestive tract, gills, and skin microbiome of Sepia officinalis (Lutz et al., 2019). The populations of S. oualaniensis are one of the most promising biological resources in the South China Sea because of their short life cycles (Zhang et al., 2010). The microbiological characteristics of the medium forms of different sexes and gonadal maturities are poorly understood. We therefore selected samples of females and males with mature and immature gonads of medium-form squid populations in this study. The differences in the intestinal and gill bacterial communities of the squid and the relationship between the bacterial community and the different sexes and gonadal maturities were analyzed. The results would augment the knowledge on the biology of squid resources in the South China Sea and enable their rational development and utilization.
Mycoplasma, belonging to Tenericutes, was the most dominant group of bacteria in the intestinal samples of FN, MN, and MY in S. oualaniensis. Mycoplasma lack cell wall (Hines et al., 2020). Different species of Mycoplasma colonize different tissues, such as the intestinal tract (Ransom, 2008) and gills (Kirchhoff & Rosengarten, 1984). Mycoplasma spp. are pathogens responsible for some human respiratory diseases (Razin, 1967), while they play a symbiotic role in the intestines of various fish (Huyben et al., 2018; Mora-Sánchez et al., 2020; Hines et al., 2020). Bano et al. (2007) found that Mycoplasma mobile could colonize the intestine of fish without causing disease. Kang et al. (2022) suspected that cephalopods may also have symbiotic relationships with gut Mycoplasma through ammonia metabolism, as in the case of salmonoids, because cephalopods are both carnivorous and ammonotelic. The intestinal bacterial community in FYI differed from that of the other types of squids, with Proteobacteria, Firmicutes, Bacteroidetes, and Tenericutes being the dominant phyla; however, Tenericutes was the dominant phylum in the FNI, MNI, and MYI groups. LEfSe analysis indicated that 26 bacterial taxa were enriched in the FYI group. In contrast, zero, seven, and nine bacterial groups were enriched in the FNI, MNI, and MYI groups, respectively. The phylum Proteobacteria (mainly members of the class Gammaproteobacteria) was one of the most abundant groups, and its members are found in various environments (Etyemez and Balcázar, 2015). The BD1-7 clade was one of the dominant bacteria in the intestines of mature male and female squid, but not in immature individuals. It was the most predominant bacteria in the gill tissue.
The microbial community structure in the gills of squid populations is different from that in the intestinal flora. The gill is an essential organ for exchange of material with the environment; therefore, the structure of the bacterial community in the gill is similar to that of the aquatic environment (Kuang et al., 2020). In this study, Proteobacteria, Firmicutes, and Bacteroidetes were the dominant phyla in gill samples of S. oualaniensis, regardless of sex and gonadal maturity. The dominant genus was the BD1-7 clade, with a relative abundance of 72.22%–76.12% in the FY, MN, and MY groups. Its relative abundance in the FN group was slightly lower (48.41%). The BD1-7 clade belonged to the oligotrophic marine Gammaproteobacteria (OMG) group (Cho and Giovannoni, 2004), which is a dominant bacterial group in marine environments, and was associated with the gorgonians of the Gorgoniidae family, suggesting that it is crucial for the health of these holobionts (van de Water et al., 2018; Yu et al., 2021). In addition, Lactobacillus and Bacteroidales S24-7 group were present in the microbial communities of the gills and intestines of squid. Zhang et al. (2022) reported that Lactobacillus and Bacteroidales S24-7 group are the intestinal probiotic for fish and shrimp, and that they are a part of the normal intestinal bacterial community (Mohammadian et al., 2016).
Relationship of the intestinal bacterial community with squid growth characteristics
Zhang P et al. (2015) found that, in mid-sized S. oualaniensis, the male: female ratio was similar in the 70−100 mm mantle length group, whereas there were more males than females in the 110−130 mm mantle length group. There were significantly more females than males in the 140 mm mantle length group. Squid with a mantle length greater than 160 mm were all females. Females reached sexual maturity when the mantle length was at least 179.7 mm, while in males it was 125.6 mm. In this study, the average mantle lengths of the FY, MY, FN, and MN groups were 190.8, 143.7, 119.5, and 112.5 mm, respectively. These results are similar to those reported by Zhang P et al. (2015). Gong et al. (2016) found that other cephalopods and fish were the main prey for the S. oualaniensis in the South China Sea. In particular, squids with more extended mantle lengths prefer to feed on larger cephalopods and fish.
Correlation analysis showed that the mantle length of S. oualaniensis was significantly negatively (p<0.05) correlated with the relative abundance of Tenericutes, but significantly positively (p<0.01) correlated with that of Firmicutes and Bacteroidetes. The mantle length and body mass results showed that the growth characteristics of squid of different sexes and maturities were different. The mantle length of FY was significantly longer than that of the other types (p<0.05). Furthermore, the intestinal bacterial communities of FYI were different from that of the other types of squids, where the relative abundance of Mycoplasma was significantly (p<0.05) lower than that in MNI, MYI, and FNI. It appears that female sexual maturity of S. oualaniensis is dependent on the consumption of small squids. With the increase in mantle length of S. oualaniensis, the number of smaller prey groups decreased, whereas that of larger ones increased. This is in agreement with the “optimal feeding theory”. Predators will prey on sizeable individual bait organisms as much as possible to maximize energy, because more energy can be obtained from large organisms (Gong et al., 2015). Kang et al. (2022) found that host body weight is associated with cephalopod gut bacterial abundance. Mycoplasma abundance showed a significant correlation with host body weight. They considered that the cephalopod diet may be correlated with the abundance of gut Mycoplasma. Mycoplasma actively metabolize ammonia in the intestine (Rasmussen et al., 2021); therefore, larger cephalopods were likely to produce more ammonia. However, based on the results of this study, we believe that FY was a unique form of S. oualaniensis, as it is different from other groups in terms of predation and reproduction, among others. The mantle length of S. oualaniensis was significantly positively correlated with the relative abundances of Lactobacillus and Bacteroidales S24-7 (p<0.05). Lactobacillus, as a probiotic, can efficiently improve growth performance, digestive enzyme activities, and intestinal microbiota (Gupta et al., 2018). The beneficial bacteria, such as Lactobacillus and Bacteroidales S24-7 group, may be better suited for the growth of S. oualaniensis in the FY group. Therefore, the differences in the diversity of intestinal bacterial community of S. oualaniensis of different sexes and maturities could be related to their growth and feeding characteristics.
Microbial metabolic function
Intestinal and gill bacterial metabolic functions could influence the stability of animal health and the aquatic environment. In this study, some bacterial metabolic processes were significantly different between the gill and intestinal samples, according to random forest KEGG analysis. In particular, the gill samples had significantly more pathways in the top 20 KEGG pathways than those of the intestinal samples (p<0.001). Among the top 20 pathways, the bacteria were mainly related to the degradation of environmental organic matter and fatty acid metabolism. This could be attributed to the contact between the bacteria in the gill tissue and water to obtain sufficient oxygen, resulting in higher metabolic activity than that in the intestinal bacteria. Zhou et al. (2019) suggested that multiple interacting bacterial species in a microbial community form a complex ecological network. Positive correlations between bacteria may represent a cooperative or complementary relationship, while negative correlations represent predatory or competitive relationships (Yang et al., 2015; Duan et al., 2020). In this study, except for OTU1 and OTU2, the other bacteria showed positive interactions. The number of positive correlations was greater than the negative correlations, indicating that the gill and intestinal bacterial communities of S. oualaniensis may have more positive interactions based on the bacterial correlation network.
Conclusion
This study demonstrated that the diversity of the intestinal bacterial community was related to the growth and feeding characteristics of S. oualaniensis of different sexes and maturities. Mycoplasma, belonging to Tenericutes, was the most dominant group of bacteria in the intestinal microbial samples of FN, MN, and MY in the S. oualaniensis populations. The intestinal microbial community structure of the FY group differed from that of other squids, with the dominant bacteria belonging to Proteobacteria, Firmicutes, Bacteroidetes, and Tenericutes. The mantle length of FY was significantly longer than that of the other types. The microbial community in the gills of the squid populations differed from that in the intestines. Proteobacteria, Firmicutes, and Bacteroidetes were the dominant phyla in the gill samples of S. oualaniensis, regardless of sex and gonadal maturity. This study provides insight into the relationship between the bacterial community of S. oualaniensis of different sexes and maturities and its growth and feeding characteristics.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA822533.
Ethics statement
The animal study was reviewed and approved by The Animal Care and Use Committee of the South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences.
Author contributions
HX drafted the manuscript. HX, SH, and WG performed the experiments. ZP and LJ collected the samples. CZ and CY conceived and designed the experiments. HX, XY, and XW analyzed the data. All authors have contributed to the manuscript and approved the submitted version.
Funding
This study was funded by the Ministry of Agriculture and Rural Affairs, P. R. China (NFZX2021); the Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2020TD54); the earmarked fund for CARS-48; the Major Projects of Basic and Applied Basic Research Programs in Guangdong Province (2019B030302004); and the Central Public-Interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (2021SD08).
Acknowledgments
We would like to thank Mingke Biotechnology Co. Ltd. (Hangzhou) for sequencing analysis and Editage (www.editage.cn) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amato K. R., Yeoman C. J., Kent A., Righini N., Carbonero F., Estrada A., et al. (2013). Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 7, 1344–1353. doi: 10.1038/ismej.2013.16
Bano N., deRae Smith A., Bennett W., Vasquez L., Hollibaugh J. T. (2007). Dominance of mycoplasma in the guts of the long-jawed mudsucker, gillichthys mirabilis, from five California salt marshes. Environ. Microbiol. 9 (10), 2636–2641. doi: 10.1111/j.14622920.2007.01381.x
Chen G. B., Zhang J., Yu J., Fan J. T., Fang L. C. (2013). Hydroacoustic scattering characteristics and biomass assessment of the purpleback flying squid [Sthenoteuthis oualaniensis, (Lesson 1830)] from the deepwater area of the south China Sea. J. Appl. Ichthyol. 29, 1447–1452. doi: 10.1111/jai.12360
Cho J. C., Giovannoni S. J. (2004). Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. . Appl. Environ. Microbiol 70 (1), 432–440. doi: 10.1128/AEM.70.1.432-440.2004
Duan Y. F., Huang J. H., Wang Y., Zhang J. S. (2020). Characterization of bacterial community in intestinal and rearing water of Penaeus monodon differing growth performances in outdoor and indoor ponds. Aquac. Res. 51, 4279–4289. doi: 10.1111/are.14770
Etyemez M., Balcázar J. L. (2015). Bacterial community structure in the intestinal ecosystem of rainbow trout (Oncorhynchus mykiss) as revealed by pyrosequencing-based analysis of 16S rRNA genes. Res. Vet. Sci. 100, 8–11. doi: 10.1016/j.rvsc.2015.03.026
Fouts D. E., Szpakowski S., Purushe J., Torralba M., Waterman R. C., MacNeil M. D., et al. (2012). Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS One 7 (11), e48289. doi: 10.1371/journal.pone.0048289
Gong Y. Y., Chen Z. Z., Zhang J., Jiang Y. E. (2015). Feeding habits of Diaphus chrysorhynchus from continental slope region in northern south Sea in autumn. South China Fish. Sci. 11 (5), 90–99. doi: 10.3969/j.issn.2095-0780.2015.05.011
Gong Y. Y., Kong X. L., Yang Y. T., Zhan F. P., Zhang P., Jiang Y. E., et al. (2018). Feeding habits of dwarf-form Sthenoteuthis oualaniensis population in the south China Sea. Mar. Fish. 40 (4), 395–403. doi: 10.13233/j.cnki.mar.fish.2018.04.002
Gong Y. Y., Zhan F. P., Yang Y. T., Zhang P., Kong X. L., Jiang Y. E., et al. (2016). Feeding habits of Symplectoteuthis oualaniensis in the south China Sea. South China Fish. Sci. 12 (4), 80–87. doi: 10.3969/j.issn.2095-0780.2016.04.010
Gupta S., Fečkaninová A., Lokesh J., Koščová J., Sørensen M., Fernandes J., et al. (2018). Lactobacillus dominate in the intestine of atlantic salmon fed dietary probiotics. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03247
Hines I. S., Ferguson C. S., Bushman T. J., Gatlin D. M., Jensen R. V., Smith S. A., et al. (2020). Impact of a yeast-based dietary supplement on the intestinal microbiome of rainbow trout, Oncorhynchus mykiss. aquac. Res. 52, 1594–1604. doi: 10.1111/are.15011
Huang H., Wei Y., Yang X. Q., Zhao Y. Q., Chen S. J., Hu X., et al. (2020). Effect of muscle modification on the quality of squid (Symplectoteuthis oualaniensis). Food Ferment. Ind. 46 (22), 42–47. doi: 10.13995/j.cnki.11-1802/ts.024676
Huyben D., Sun L., Moccia R., Kiessling A., Dicksved J., Lundh T. (2018). Dietary live yeast and increased water temperature influence the gut microbiota of rainbow trout. J. Appl. Microbiol. 124 (6), 1377–1392. doi: 10.1111/jam.13738
Iehata S., Valenzuela F., Riquelme C. (2015). Analysis of bacterial community and bacterial nutritional enzyme activity associated with the digestive tract of wild Chilean octopus (Octopus mimus Gould 1852). Aquac. Res. 46 (4), 861–873. doi: 10.1111/are.12240
Jiang Y. E., Zhang P., Lin Z. J., Qiu Y. S., Fang Z. Q., Chen Z. Z. (2015). Statolith morphology of purpleback flying squid (Sthenoeuthis oualaniensis) in the offshore south China Sea. South China Fish. Sci. 11 (5), 27–37. doi: 10.3969/j.issn.2095-0780.2015.05.004
Jin Y. X., Wu S. S., Zeng Z. Y., Fu Z. W. (2017). Effects of environmental pollutants on gut microbiota. Environ. pollut. 222, 1–9. doi: 10.1016/j.envpol.2016.11.045
Kang W., Kim P. S., Tak E. J., Sung H., Shin N. R., Hyun D. W., et al. (2022). Host phylogeny, habitat, and diet are main drivers of the cephalopod and mollusk gut microbiome. Anim. Microbiome. 4, 30. doi: 10.1186/s42523-022-00184-x
Kirchhoff H., Rosengarten R. (1984). Isolation of a motile mycoplasma from fish. J. Gen. Microbiol. 130 (9), 2439–2445. doi: 10.1099/00221287-130-9-2439
Kuang T. X., He A. Y., Lin Y. F., Huang X. D., Liu L., Zhou L. (2020). Comparative analysis of microbial communities associated with the gill, gut, and habitat of two filter-feeding fish. Aquac. Rep. 18, 100501. doi: 10.1016/j.aqrep.2020.100501
Li Y. M., Zhang K., Liu Y., Li K., Hu D. F., Wronski T. (2019). Community composition and diversity of intestinal microbiota in captive and reintroduced przewalski’s horse (Equus ferus przewalskii). Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01821
Li M., Zhang P., Zhang J., Zhang K., Chen Z. Z. (2019). Genetic differentiation of the purpleback flying squid, Sthenoteuthis oualaniensis, in the south China Sea: population or species divergence. J. Fish. Sci. China 26 (1), 133–140. doi: 10.3724/SP.J.1118.2019.18193
Lutz H. L., Ramírez-Puebla S. T., Abbo L., Durand A., Schlundt C., Gottel N. R., et al. (2019). A simple microbiome in the European common cuttlefish, Sepia officinalis. mSystems. 4 (4), e177–e119. doi: 10.1128/mSystems.00177-19
Mohammadian T, Alishahi M, Tabandeh M. R, Ghorbanpoor M, Gharibi D, Tollabi M, et al (2016). Probiotic effects of Lactobacillus plantarum and L. delbrueckii ssp. bulguricus on some immune-related parameters in Barbus grypus. Aquac. Int. 24, 225–242. doi: 10.1007/s10499-015-9921-8
Mora-Sánchez B., Pérez-Sánchez T., Balcázar J. L. (2020). Phylogenetic analysis of intestinal microbiota reveals novel Mycoplasma phylotypes in salmonid species. Microb. Pathog. 145, 104210. doi: 10.1016/j.micpath.2020.104210
Paradis E., Claude J., Strimmer K. (2004). APE: Analyses of phylogenetics and evolution in r language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Ransom B. L. (2008). Intestinal microbial community composition of six actinopterygii fish species in the southeastern united states (Athens, GA: University of Georgia).
Rasmussen J. A., Villumsen K. R., Duchêne D. A., Puetz L. C., Delmont T. O., Sveier H., et al. (2021). Genome-resolved metagenomics suggests a mutualistic relationship between Mycoplasma and salmonid hosts. Commun. Biol. 4 (1), 1–10. doi: 10.1038/s42003-021-02105-1
Schloss P. D., Gevers D., Westcott S. L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6, e27310. doi: 10.1371/journal.pone.0027310
Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Strugnell J., Nishiguchi M. K. (2007). Molecular phylogeny of coleoid cephalopods (Mollusca: Cephalopoda) inferred from three mitochondrial and six nuclear loci: a comparison of alignment, implied alignment and analysis methods. J. Molluscan Stud. 73 (4), 399–410. doi: 10.1093/mollus/eym038
Sun F. L., Xia X. M., Simon M., Wang Y. S., Zhao H., Sun C. C., et al. (2022). Anticyclonic eddy driving significant changes in prokaryotic and eukaryotic communities in the south China Sea. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.773548
van de Water J. A. J. M., Voolstra C. R., Rottier C., Cocito S., Peirano A., Allemand D., et al. (2018). Seasonal stability in the microbiomes of temperate gorgonians and the red coral Corallium rubrum across the mediterranean sea. Microb. Ecol. 75, 274–288. doi: 10.1007/s00248-017-1006-y
Wang R. G., Chen X. J. (2005). World oceanic economic ommastrephidae resources and their fisheries (Beijing: Ocean Press), 284–296.
Wang A. R., Ran C., Ringø E., Zhou Z. G. (2018). Progress in fish gastrointestinal microbiota research. Rev. Aquac. 10 (3), 626–640. doi: 10.1111/raq.12191
Webster N. S., Taylor M. W., Behnam F., Lücker S., Rattei T., Whalan S., et al. (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12 (8), 2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x
Yang G., Xu Z., Tian X. L., Dong S. L., Peng M. (2015). Intestinal microbiota and immune related genes in sea cucumber (Apostichopus japonicus) response to dietary β-glucan supplementation. Biochem. Biophys. Res. Commun. 458, 98–103. doi: 10.1016/j.bbrc.2015.01.074
Yu X. P., Yu K. F., Liao Z. H., Chen B., Deng C. Q., Yu J. Y., et al. (2021). Seasonal fluctuations in symbiotic bacteria and their role in environmental adaptation of the scleractinian coral Acropora pruinosa in high-latitude coral reef area of the south China Sea. Sci. Total Environ. 792, 148438. doi: 10.1016/j.scitotenv.2021.148438
Zhang J., Chen Z. Z., Chen G. B., Zhang P., Qiu Y. S., Yao Z. (2015). Hydroacoustic studies on the commercially important squid Sthenoteuthis oualaniensis in the south China Sea. Fish. Res. 169, 45–51. doi: 10.1016/j.fishres.2015.05.003
Zhang X., Li M., Tao X. Y., Yang Y. H., Sun P., Jin M., et al. (2022). Effects of dietary montmorillonite supplementation on the growth performance, antioxidant capacity, intestinal barrier and microbiota composition in Marsupenaeus japonicus. Aquaculture. 557, 738330. doi: 10.1016/j.aquaculture.2022.738330
Zhang P., Yang L., Zhang X. F., Tang Y. G. (2010). The present status and prospect on exploitation of tuna and squid fishery resources in south China Sea. South China Fish. Sci. 6 (1), 68–74. doi: 10.3969/j.issn.1673-2227.2010.01.012
Zhang P., Yan L., Yang B. Z., Tan Y. G., Zhang X. F., Chen S., et al. (2015). Population structure of purpleback flying squid (Sthenoteuthis oualaniensis) in nansha area in spring. South China Fish. Sci. 11 (5), 11–19. doi: 10.3969/j.issn.2095-0780.2015.05.002
Zhou L. I., Chen C., Xie J., Xu C., Zhao Q., Qin J. G., et al. (2019). Intestinal bacterial signatures of the “cotton shrimp-like” disease explain the change of growth performance and immune responses in pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol. 92, 629–636. doi: 10.1016/j.fsi.2019.06.054
Zhu K., Wang X. H., Zhang P., Du F. Y., Qiu Y. S. (2019). A study on morphological indicators variations and discrimination of medium and dwarf forms of purple flying squid Sthenoteuthis oualaniensis in the central and southern south China Sea. Periodical Ocean Univ. China. 49 (1), 43–54. doi: 10.16441/j.cnki.hdxb.20170251
Keywords: microbial community structure, Sthenoteuthis oualaniensis, medium-form populations, different sexes and gonadal maturities, South China Sea
Citation: Hu X, Su H, Zhang P, Chen Z, Xu Y, Xu W, Li J, Wen G and Cao Y (2022) Microbial community characteristics of the intestine and gills of medium-form populations of Sthenoteuthis oualaniensis in the South China Sea. Front. Mar. Sci. 9:920536. doi: 10.3389/fmars.2022.920536
Received: 14 April 2022; Accepted: 25 July 2022;
Published: 12 August 2022.
Edited by:
Piotr Rzymski, Poznan University of Medical Sciences, PolandReviewed by:
Carmen Rizzo, Stazione Zoologica Anton Dohrn Napoli, ItalyYan Fang, Ludong University, China
Copyright © 2022 Hu, Su, Zhang, Chen, Xu, Xu, Li, Wen and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yucheng Cao, cyc_715@163.com
 Xiaojuan Hu1,2,3
Xiaojuan Hu1,2,3  Zuozhi Chen
Zuozhi Chen Yucheng Cao
Yucheng Cao