Abstract
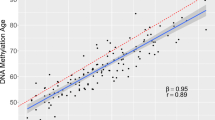
DNA methylation (DNAm) patterns across the genome changes during aging and development of complex diseases including type 2 diabetes (T2D). Our study aimed to estimate DNAm trajectories of CpG sites associated with T2D, epigenetic age (DNAmAge), and age acceleration based on four epigenetic clocks (GrimAge, Hannum, Horvath, phenoAge) in the period 10 years prior to and up to T2D onset. In this nested case–control study within Doetinchem Cohort Study, we included 132 incident T2D cases and 132 age- and sex-matched controls. DNAm was measured in blood using the Illumina Infinium Methylation EPIC array. From 107 CpG sites associated with T2D, 10 CpG sites (9%) showed different slopes of DNAm trajectories over time (p < 0.05) and an additional 8 CpG sites (8%) showed significant differences in DNAm levels (at least 1%, p-value per time point < 0.05) at all three time points with nearly parallel trajectories between incident T2D cases and controls. In controls, age acceleration levels were negative (slower epigenetic aging), while in incident T2D cases, levels were positive, suggesting accelerated aging in the case group. We showed that DNAm levels at specific CpG sites, up to 10 years before T2D onset, are different between incident T2D cases and healthy controls and distinct patterns of clinical traits over time may have an impact on those DNAm profiles. Up to 10 years before T2D diagnosis, cases manifested accelerated epigenetic aging. Markers of biological aging including age acceleration estimates based on Horvath need further investigation to assess their utility for predicting age-related diseases including T2D.




Similar content being viewed by others
References
Saeedi P, Petersohn I, Salpea P, et al Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Research and Clinical Practice. 2019;157. https://doi.org/10.1016/j.diabres.2019.107843
Willemsen G, Ward KJ, Bell CG, et al. The concordance and heritability of type 2 diabetes in 34,166 twin pairs from International Twin Registers: The Discordant Twin (DISCOTWIN) Consortium. Twin Res Hum Genet. 2015;18(6):762–71. https://doi.org/10.1017/thg.2015.83.
Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–13. https://doi.org/10.1038/s41588-018-0241-6.
Xue A, Wu Y, Zhu Z, et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun. 2018;9(1):2941. https://doi.org/10.1038/s41467-018-04951-w.
Gill JMR, Cooper AR. Physical activity and prevention of type 2 diabetes mellitus. Sports Med. 2008;38(10):807–24. https://doi.org/10.2165/00007256-200838100-00002.
Carter P, Gray LJ, Troughton J, Khunti K, Davies MJ. Fruit and vegetable intake and incidence of type 2 diabetes mellitus: systematic review and meta-analysis. BMJ. 2010;341: c4229. https://doi.org/10.1136/bmj.c4229.
Franks PW, Pearson E, Florez JC. Gene-environment and gene-treatment interactions in type diabetes. Diabetes Care. 2013;36(5):1413–21. https://doi.org/10.2337/dc12-2211.
Liu L, Li Y, Tollefsbol TO. Gene-environment interactions and epigenetic basis of human diseases. Curr Issues Mol Biol. 2008;10(1–2):25–36.
Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21.
Jin Z, Liu Y. DNA methylation in human diseases. Genes and Diseases. 2018;5:1–8.
Cardona A, Day FR, Perry JRB, et al Epigenome-wide association study of incident type 2 diabetes in a British population: EPIC-Norfolk study. Diabetes db180290. 2019;
Wahl S, Drong A, Lehne B, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–6. https://doi.org/10.1038/nature20784.
2. Classification and diagnosis of diabetes: <em>Standards of Medical Care in Diabetes—2019</em> Diabetes Care 42(Supplement 1) 2019;S13 LP-S28. https://doi.org/10.2337/dc19-S002
Lowsky DJ, Olshansky SJ, Bhattacharya J, Goldman DP. Heterogeneity in healthy aging. The Journals of Gerontology: Series A. 2013;69(6):640–9. https://doi.org/10.1093/gerona/glt162.
Jylhävä J, Pedersen NL, Hägg S. Biological age predictors EBioMedicine. 2017;21:29–36.
Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. https://doi.org/10.1186/gb-2013-14-10-r115.
Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11(2):303–27. https://doi.org/10.18632/aging.101684.
Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–67. https://doi.org/10.1016/j.molcel.2012.10.016.
Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–91. https://doi.org/10.18632/aging.101414.
Hillary RF, Stevenson AJ, McCartney DL, et al. Epigenetic measures of ageing predict the prevalence and incidence of leading causes of death and disease burden. Clin Epigenetics. 2020;12(1):115. https://doi.org/10.1186/s13148-020-00905-6.
Grant CD, Jafari N, Hou L, et al. A longitudinal study of DNA methylation as a potential mediator of age-related diabetes risk. Geroscience. 2017;39(5–6):475–89. https://doi.org/10.1007/s11357-017-0001-z.
Walaszczyk E, Luijten M, Spijkerman AMW, et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case–control sample of the Lifelines study. Diabetologia. 2018;61(2):354–68. https://doi.org/10.1007/s00125-017-4497-7.
Fraszczyk E, Spijkerman AMW, Zhang Y, et al. Epigenome-wide association study of incident type 2 diabetes: a meta-analysis of five prospective European cohorts. Diabetologia. 2022;65(5):763–76. https://doi.org/10.1007/s00125-022-05652-2.
Verschuren W, Blokstra A, Picavet H, Smit H. Cohort profile: the Doetinchem Cohort Study. Int J Epidemiol. 2008;37(6):1236–41. https://doi.org/10.1093/ije/dym292.
Picavet HSJ, Blokstra A, Spijkerman AMW, Verschuren WMM. Cohort profile update: the Doetinchem Cohort Study 1987–2017: lifestyle, health and chronic diseases in a life course and ageing perspective. Int J Epidemiol. 2017;46(6):1751–1751g. https://doi.org/10.1093/ije/dyx103.
WHO Classification of diabetes mellitus. https://apps.who.int/iris/handle/10665/325182
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215.
Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9. https://doi.org/10.1093/bioinformatics/btu049.
Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–e47. https://doi.org/10.1093/nar/gkv007.
Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. https://doi.org/10.1093/biostatistics/kxj037.
Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. https://doi.org/10.1186/1471-2105-13-86.
Sharp GC, Arathimos R, Reese SE, et al. Maternal alcohol consumption and offspring DNA methylation: findings from six general population-based birth cohorts. Epigenomics. 2018;10(1):27–42. https://doi.org/10.2217/epi-2017-0095.
DNA methylation age and the epigenetic clock. http://labs.genetics.ucla.edu/horvath/htdocs/dnamage/
Kulkarni H, Kos MZ, Neary J, et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet. 2015;24(18):5330–44. https://doi.org/10.1093/hmg/ddv232.
Chambers JC, Loh M, Lehne B, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3(7):526–34. https://doi.org/10.1016/S2213-8587(15)00127-8.
Al Muftah WA, Al-Shafai M, Zaghlool SB, et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin Epigenetics. 2016;8:13. https://doi.org/10.1186/s13148-016-0177-6.
Pinheiro JC, Bates DJ, DebRoy S, Sakar D The Nlme package: linear and nonlinear mixed effects models, R Version. 2012;3
Li M, Zou D, Li Z, et al. EWAS Atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019;47(D1):D983–8. https://doi.org/10.1093/nar/gky1027.
Kriebel J, Herder C, Rathmann W, et al. Association between DNA methylation in whole blood and measures of glucose metabolism: KORA F4 study. PLoS ONE. 2016;11(3): e0152314. https://doi.org/10.1371/journal.pone.0152314.
Soriano-Tárraga C, Jiménez-Conde J, Giralt-Steinhauer E, et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum Mol Genet. 2016;25(3):609–19. https://doi.org/10.1093/hmg/ddv493.
Florath I, Butterbach K, Heiss J, et al. Type 2 diabetes and leucocyte DNA methylation: an epigenome-wide association study in over 1,500 older adults. Diabetologia. 2016;59(1):130–8. https://doi.org/10.1007/s00125-015-3773-7.
Duran J, Obach M, Navarro-Sabate A, et al. Pfkfb3 is transcriptionally upregulated in diabetic mouse liver through proliferative signals. The FEBS Journal. 2009;276(16):4555–68. https://doi.org/10.1111/j.1742-4658.2009.07161.x.
Tobi EW, Slieker RC, Luijk R, et al DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Science Advances. 2018;4(1). https://doi.org/10.1126/sciadv.aao4364
Stover PJ, Field MS. Vitamin B-6. Adv Nutr. 2015;6(1):132–3. https://doi.org/10.3945/an.113.005207.
Mascolo E, Vernì F Vitamin B6 and diabetes: relationship and molecular mechanisms. International Journal of Molecular Sciences. 2020;21(10). https://doi.org/10.3390/ijms21103669
Nix WA, Zirwes R, Bangert V, et al. Vitamin B status in patients with type 2 diabetes mellitus with and without incipient nephropathy. Diabetes Research and Clinical Practice. 2015;107(1):157–65. https://doi.org/10.1016/j.diabres.2014.09.058.
Ahn HJ, Min KW, Cho Y-O. Assessment of vitamin B(6) status in Korean patients with newly diagnosed type 2 diabetes. Nutr Res Pract. 2011;5(1):34–9. https://doi.org/10.4162/nrp.2011.5.1.34.
Satyanarayana A, Balakrishna N, Pitla S, et al. Status of B-vitamins and homocysteine in diabetic retinopathy: association with vitamin-B12 deficiency and hyperhomocysteinemia. PLOS ONE. 2011;6(11):e26747.
Ligthart S, Marzi C, Aslibekyan S, et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016;17(1):255. https://doi.org/10.1186/s13059-016-1119-5.
del Gomez-Alonso M, C, Kretschmer A, Wilson R, et al. DNA methylation and lipid metabolism: an EWAS of 226 metabolic measures. Clin Epigenetics. 2021;13(1):7. https://doi.org/10.1186/s13148-020-00957-8.
Truong V, Huang S, Dennis J, et al. Blood triglyceride levels are associated with DNA methylation at the serine metabolism gene PHGDH. Sci Rep. 2017;7(1):11207. https://doi.org/10.1038/s41598-017-09552-z.
Hedman ÅK, Mendelson MM, Marioni RE, et al. Epigenetic patterns in blood associated with lipid traits predict incident coronary heart disease events and are enriched for results from genome-wide association studies. Circ Cardiovasc Genet. 2017;10(1): e001487. https://doi.org/10.1161/CIRCGENETICS.116.001487.
Mendelson MM, Marioni RE, Joehanes R, et al. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Med. 2017;14(1): e1002215.
Eberlé D, Clément K, Meyre D, et al. SREBF-1 gene polymorphisms are associated with obesity and type 2 diabetes in French obese and diabetic cohorts. Diabetes. 2004;53(8):2153–7. https://doi.org/10.2337/diabetes.53.8.2153.
Ali O, Cerjak D, Kent JW Jr, et al. Methylation of SOCS3 is inversely associated with metabolic syndrome in an epigenome-wide association study of obesity. Epigenetics. 2016;11(9):699–707. https://doi.org/10.1080/15592294.2016.1216284.
Bacos K, Gillberg L, Volkov P, et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun. 2016;7:11089. https://doi.org/10.1038/ncomms11089.
Rönn T, Volkov P, Gillberg L, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24(13):3792–813. https://doi.org/10.1093/hmg/ddv124.
Field AE, Robertson NA, Wang T, Havas A, Ideker T, Adams PD. DNA methylation clocks in aging: categories, causes, and consequences. Mol Cell. 2018;71:882–95.
Li X, Ploner A, Wang Y, et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. Elife. 2020;9: e51507. https://doi.org/10.7554/eLife.51507.
el Khoury LY, Gorrie-Stone T, Smart M, et al. Systematic underestimation of the epigenetic clock and age acceleration in older subjects. Genome Biol. 2019;20(1):283. https://doi.org/10.1186/s13059-019-1810-4.
Lee K, Pausova Z Cigarette smoking and DNA methylation. Frontiers in Genetics. 2013;4
Marioni RE, Shah S, McRae AF, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16(1):25. https://doi.org/10.1186/s13059-015-0584-6.
Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics. 2016;8(1):64. https://doi.org/10.1186/s13148-016-0228-z.
Zheng Y, Joyce BT, Colicino E, et al. Blood epigenetic age may predict cancer incidence and mortality. EBioMedicine. 2016;5:68–73. https://doi.org/10.1016/j.ebiom.2016.02.008.
Joyce BT, Gao T, Zheng Y, et al. Epigenetic age acceleration reflects long-term cardiovascular health. Circ Res. 2021;129(8):770–81. https://doi.org/10.1161/CIRCRESAHA.121.318965.
Hillary RF, Stevenson AJ, Cox SR, et al. An epigenetic predictor of death captures multi-modal measures of brain health. Mol Psychiatry. 2021;26(8):3806–16. https://doi.org/10.1038/s41380-019-0616-9.
Verschoor CP, Lin DTS, Kobor MS, et al. Epigenetic age is associated with baseline and 3-year change in frailty in the Canadian Longitudinal Study on Aging. Clin Epigenetics. 2021;13(1):163. https://doi.org/10.1186/s13148-021-01150-1.
Acknowledgements
We thank the participants of the Doetinchem Cohort Study, as well as the field workers of the Municipal Health Services in Doetinchem (C. te Boekhorst, I. Hengeveld, L. de Klerk, I. Thus, and C. de Rover) for their contribution to the data collection of this study. We are grateful to P. Vissink for logistic management and A. Blokstra for data management (both from the National Institute for Public Health and the Environment).
Funding
This work was supported by the Ministry of Health, Welfare and Sport of the Netherlands, the National Institute for Public Health and the Environment (RIVM; grant number S/132005), and by Biobanking and Biomolecular Resources Research Infrastructure-NL (grant number CP2011-27).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Fraszczyk, E., Thio, C.H.L., Wackers, P. et al. DNA methylation trajectories and accelerated epigenetic aging in incident type 2 diabetes. GeroScience 44, 2671–2684 (2022). https://doi.org/10.1007/s11357-022-00626-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-022-00626-z