Abstract
Multicellularity was a watershed development in evolution. However, it also meant that individual cells could escape regulatory mechanisms that restrict proliferation at a severe cost to the organism: cancer. From the standpoint of cellular organization, evolutionary complexity scales to organize different molecules within the intracellular milieu. The recent realization that many biomolecules can “phase-separate” into membraneless organelles, reorganizing cellular biochemistry in space and time, has led to an explosion of research activity in this area. In this review, we explore mechanistic connections between phase separation and cancer-associated processes and emerging examples of how these become deranged in malignancy.
One of the fundamental functions of phase separation is to rapidly and dynamically respond to environmental perturbations. Importantly, these changes often lead to alterations in cancer-relevant pathways and processes. This review covers recent advances in the field, including emerging principles and mechanisms of phase separation in cancer.
INTRODUCTION
Malignant transformation is exemplified by the breakdown of regulatory processes that keep the proliferation of postmitotic cells under control. Key among these are maintenance of genomic integrity, regulation of gene expression, and cell signaling. Maintenance of proper subcellular localization is critical for these processes, and the roles of organelles such as the nucleus, mitochondria, and the Golgi apparatus in these functions have been investigated for decades. Yet it has recently been appreciated that subcellular organization includes myriad organelle-like structures that are not membrane bound. Many biomolecules can undergo a process known as liquid–liquid phase separation (LLPS), which can sequester specific proteins and nucleic acids into membraneless organelles (MLO), reorganizing cellular biochemistry in space and time (Fig. 1A). The past decade has featured an explosion of research activity in this area, and many excellent reviews have summarized its mechanistic underpinnings and roles in development and neurologic diseases (1–6). By contrast, the exploration of these concepts in cancer has begun more recently (1, 7–13). In this review, we present evidence that known cancer drivers are enriched in genes that encode proteins that undergo phase separation (Fig. 1B). Furthermore, we explore mechanistic connections between phase separation and cancer-associated processes and how these can become deranged in disease. Finally, we discuss how LLPS may provide new therapeutic opportunities in oncology.
An emerging field in LLPS in cancer. A, Biomolecular polymers, such as proteins with intrinsically disordered regions, can undergo phase transitions resulting in liquid droplets with higher local density of the protein and physical properties distinct from the surroundings. Over time, the liquid droplets can mature into other phase-separated species such as gels, oligomers, or fibers. B, Increasing appreciation in studies over the last two decades exploring the connections between LLPS and cancer. Cyan bars represent the number of publications found in PubMed related to LLPS, which has steadily but continuously increased since 2000. Blue bars show a tremendous increase in the number of publications related to cancer and LLPS over the past 4 years. The pie chart reveals that only about 10% of LLPS genes annotated as cancer genes by the Cancer Gene Census have been studied to understand the molecular mechanisms behind cancer mutations.
An emerging field in LLPS in cancer. A, Biomolecular polymers, such as proteins with intrinsically disordered regions, can undergo phase transitions resulting in liquid droplets with higher local density of the protein and physical properties distinct from the surroundings. Over time, the liquid droplets can mature into other phase-separated species such as gels, oligomers, or fibers. B, Increasing appreciation in studies over the last two decades exploring the connections between LLPS and cancer. Cyan bars represent the number of publications found in PubMed related to LLPS, which has steadily but continuously increased since 2000. Blue bars show a tremendous increase in the number of publications related to cancer and LLPS over the past 4 years. The pie chart reveals that only about 10% of LLPS genes annotated as cancer genes by the Cancer Gene Census have been studied to understand the molecular mechanisms behind cancer mutations.
MECHANISMS OF PHASE SEPARATION
Phase separation refers to a phenomenon in which biological macromolecules demix into separate dilute and dense phases that are bounded by a surface and exhibit distinct physical properties (2). This phenomenon, driven by noncovalent, transient interactions between biopolymers, organizes proteins, and nucleic acids into MLOs. MLOs can manifest as liquids, gels, glasses, crystalline solids, and even multiphase bodies (e.g., a solid core enveloped by a liquid)and can be classified by the arrangement (e.g., crystalline or amorphous), density, and mobility of their constituent molecules (14, 15). MLOs form in a concentration-dependent manner. Once the concentration of a phase-separating protein reaches a critical threshold, protein–protein interactions become more thermodynamically favorable than protein–solvent interactions, and the system undergoes a phase transition into dense and dilute phases. Remarkably, in single-component phase separation, further increases in protein concentration cause the MLOs to grow in volume but retain the same concentration. Changing concentrations in multicomponent phase separation by contrast vary partition coefficients of individual components in a manner that minimizes the free energy of the condensate (16). Phase separation is often reversible and can be controlled by the posttranslational modification of constituent proteins or changes to the local physicochemical environment such as salt concentration, pH, temperature, and pressure (17). It is also possible for the nucleating protein species that initiated the MLO formation to recruit other molecules (e.g., members of multiprotein complexes, ligands, chaperones, other MLO-forming proteins), thereby creating a mixture of species in the MLO. MLOs are dynamic structures, and over time condensates can even harden into solids or evolve into multiphase bodies (refs. 15, 18; Fig. 1A).
Deciphering the biological importance of phase separation has sparked intense research interest. Multiple well-studied cellular structures are now appreciated to exist as MLOs, such as the nucleolus and stress granules (19, 20). MLOs allow cells to spatiotemporally organize metabolic pathways and signaling cascades, store biomolecules for later use, or adapt to stressful conditions (21, 22). However, a number of neurodegenerative diseases have been associated with the formation of aberrant aggregates, such as amyloid fibers, that may arise from phase-separated MLOs (4). The potential for phase-separated, membraneless organelles to participate in normal cellular processes—and the derangement of such processes typical of disease states—depends on the functions of constituent molecules, the biophysical properties of the phase-separated body, and their interaction with the proteostasis network.
A key feature of proteins that phase separate is the capacity to form multiple weak, transient, noncovalent interactions with other molecules. One way to achieve this property is via repeated binding domains within a biomolecule (typically referred to as multivalency). Many proteins with extended intrinsically disordered regions (IDR) are also capable of forming an MLO. IDRs do not have a single, stable conformation in solution but rather explore a range of conformations, which allows them to form weak, transient interactions with many binding partners. These regions are often depleted of hydrophobic residues and typically contain more polar and charged residues, but the mechanisms through which IDRs drive phase transition are still being uncovered. For example, some mechanisms involve the patterning of charged residues throughout the IDR, whereas others rely on infrequent aromatic residues for pi–pi stacking interactions. Proteins with IDRs are very common in the human proteome (∼35% of all amino acids are predicted to be disordered) and have weak sequence level conservation over evolutionary time (23). The presence of IDRs, despite their lack of primary sequence conservation, and specific physicochemical features of these regions are often maintained across large evolutionary distances in regulators of transcription, translation, and signaling (24, 25).
SYSTEMS-WIDE ENRICHMENT OF PHASE SEPARATION IN CANCER-ASSOCIATED PROTEINS
Given the enrichment for IDRs in signaling and gene regulatory functions—each biochemical processes with strong links to malignancy—we explored the 287 genes with annotated cancer hallmarks from the Catalogue of Somatic Mutations in Cancer (COSMIC) for their ability to encode proteins that can undergo phase separation (26). There are multiple annotated public databases (Table 1) available for studying LLPS that cover phase-separating proteins, MLOs, phase-separating RNA, IDRs, posttranslational modifications, and short linear motifs. Furthermore, there are many computational tools available for analyzing molecular features related to LLPS within protein, RNA, and DNA sequences (Table 1). Fifty-three COSMIC proteins are annotated to undergo LLPS in at least two databases (27–30), a strong enrichment relative to all genes with annotated biological processes from the Gene Ontology Consortium (GO; ref. 31; P < 1.41 × 10−6 by permutation test; Fig. 2A). Remarkably, comparisons with other disease processes indicate that phase-separating proteins are more enriched in cancer driver genes than any other evaluated disease process, including neurodegeneration (ref. 32; Fig. 2B). This enrichment spans the majority of cancer hallmarks, including maintenance of genomic stability (e.g., BLM, ERCC2, ATM, and ATR), aberrant proliferation signaling (e.g., MYC, ESR1, and ABL1), and regulation of gene expression (e.g., BRD4, CTCF, and SF3B1; Fig. 3A and B).
Computational resources, tools, and methods for studying LLPS
| Annotated public databases for studying LLPS . | ||||
|---|---|---|---|---|
| Database . | Entries . | Type . | Description . | Availability . |
| DrLLPS | 9,285 | Protein | Manually curated to compile known and predicted LLPS proteins. | http://llps.biocuckoo.cn/ |
| PhaSePro | 121 | Protein | Manually curated to list proteins that drive LLPS. | https://phasepro.elte.hu/ |
| LLPSDB | 273 | Protein | Manually curated to list LLPS-related proteins. | http://www.bio-comp.org.cn/llpsdb/home.html |
| PhaSepDB | 961 | Protein | Manually curated to list LLPS- and MLO-related proteins. | http://db.phasep.pro/ |
| AmyPro | 162 | Protein | Manually curated to list validated amyloid precursor proteins and prions. | https://amypro.net/#/ |
| DisProt | 932 | Protein | Manually curated to identify IDPs. | https://disprot.org/ |
| ZipperDB | 20,000+ | Protein | Collection of protein regions that have the tendency to form fibrils. | https://services.mbi.ucla.edu/zipperdb/ |
| MloDisDB | 719 | MLO | Manually curated to showcase the relation between MLOs and diseases. | http://mlodis.phasep.pro/ |
| RPS | 21,613 | RNA | Manually curated to list validated RNA involved in LLPS. | http://rps.renlab.org/#/Home |
| RNAPhaSep | 1,113 | RNA | Manually curated to compile known LLPS-related RNAs. | http://www.rnaphasep.cn/#/Home |
| PhosphoSitePlus | 484,496 | PTM | Curated to list commonly studied PTMs that were observed experimentally. | https://www.phosphosite.org/homeAction.action |
| dbPTM | 2,235,664 | PTM | Collection of experimentally verified protein PTMs. | https://awi.cuhk.edu.cn/dbPTM/ |
| MobiDB | 219,740,215 | IDR | Collection of IDRs and similar features on proteins. | https://mobidb.bio.unipd.it/ |
| LMPID | 1,762 | Motif | Manually curated to identify linear motifs that mediate protein interactions. | http://bicresources.jcbose.ac.in/ssaha4/lmpid/ |
| ELM DB | 3,934 | Motif | Manually curated to identify short linear motif instances. | http://elm.eu.org/ |
| Annotated public databases for studying LLPS . | ||||
|---|---|---|---|---|
| Database . | Entries . | Type . | Description . | Availability . |
| DrLLPS | 9,285 | Protein | Manually curated to compile known and predicted LLPS proteins. | http://llps.biocuckoo.cn/ |
| PhaSePro | 121 | Protein | Manually curated to list proteins that drive LLPS. | https://phasepro.elte.hu/ |
| LLPSDB | 273 | Protein | Manually curated to list LLPS-related proteins. | http://www.bio-comp.org.cn/llpsdb/home.html |
| PhaSepDB | 961 | Protein | Manually curated to list LLPS- and MLO-related proteins. | http://db.phasep.pro/ |
| AmyPro | 162 | Protein | Manually curated to list validated amyloid precursor proteins and prions. | https://amypro.net/#/ |
| DisProt | 932 | Protein | Manually curated to identify IDPs. | https://disprot.org/ |
| ZipperDB | 20,000+ | Protein | Collection of protein regions that have the tendency to form fibrils. | https://services.mbi.ucla.edu/zipperdb/ |
| MloDisDB | 719 | MLO | Manually curated to showcase the relation between MLOs and diseases. | http://mlodis.phasep.pro/ |
| RPS | 21,613 | RNA | Manually curated to list validated RNA involved in LLPS. | http://rps.renlab.org/#/Home |
| RNAPhaSep | 1,113 | RNA | Manually curated to compile known LLPS-related RNAs. | http://www.rnaphasep.cn/#/Home |
| PhosphoSitePlus | 484,496 | PTM | Curated to list commonly studied PTMs that were observed experimentally. | https://www.phosphosite.org/homeAction.action |
| dbPTM | 2,235,664 | PTM | Collection of experimentally verified protein PTMs. | https://awi.cuhk.edu.cn/dbPTM/ |
| MobiDB | 219,740,215 | IDR | Collection of IDRs and similar features on proteins. | https://mobidb.bio.unipd.it/ |
| LMPID | 1,762 | Motif | Manually curated to identify linear motifs that mediate protein interactions. | http://bicresources.jcbose.ac.in/ssaha4/lmpid/ |
| ELM DB | 3,934 | Motif | Manually curated to identify short linear motif instances. | http://elm.eu.org/ |
| Computational tools for analyzing molecular features related to LLPS . | ||||
|---|---|---|---|---|
| Tool . | Focus and methodology . | Availability . | ||
| PLAAC | Identifies prion-like domains in proteins using a hidden Markov model. | http://plaac.wi.mit.edu/ | ||
| IUPred3 | Identifies disordered protein regions based on an energy estimation method. | https://iupred3.elte.hu/ | ||
| PSPer | First in silico tool to identify prion-like RNA-binding phase separation proteins. | https://www.bio2byte.be/b2btools/psp/ | ||
| PSPredictor | Predicts phase separation proteins using machine learning with LLPSDB. | http://www.pkumdl.cn:8000/PSPredictor/ | ||
| PScore | Predicts phase separation proteins based on pi-interaction frequencies. | https://elifesciences.org/articles/31486 | ||
| catGRANULE | Predicts granule propensity based on multiple features like RNA binding. | http://service.tartaglialab.com/update_submission/475471/abfa5da35f | ||
| dSCOPE | Predicts key regions of proteins for LLPS using a machine learning algorithm, random forest. | http://dscope.omicsbio.info/ | ||
| BIAPSS | Provides interactive statistical analyses of LLPS proteins listed in PhaSePro and LLPSDB for inferring phase-separating affinities and determining biophysical statistics of LLPS proteins. | https://biapss.chem.iastate.edu/ | ||
| PrionW | Predicts prion-like domains and amyloid nucleating cores from protein sequences. | http://bioinf.uab.cat/prionw/ | ||
| MusiteDeep | Provides a deep-learning framework for predicting and visualizing PTMs in proteins. | https://www.musite.net/ | ||
| SLiMAN | Provides a webserver for analyzing and predicting linear motifs and interactions in proteins. | http://sliman.cbs.cnrs.fr/LIMIP/limip_index.py | ||
| ESpritz | Predicts protein disorder using bidirectional recursive neural networks. | http://old.protein.bio.unipd.it/espritz/ | ||
| PLATINUM | Analyzes hydrophobic and hydrophilic characteristics of proteins and other molecules. | https://model.nmr.ru/platinum/ | ||
| PSAP | Provides a machine learning classifier that analyzes proteins and predicts phase separation. | https://github.com/Guido497/phase-separation | ||
| CIDER | Predicts and annotates protein sequences for IDRs/IDPs for their conformational properties. | http://pappulab.wustl.edu/CIDER/ | ||
| DisMeta | Provides a meta-server that has predictors for disordered and low-complexity regions in proteins. | https://montelionelab.chem.rpi.edu/dismeta/ | ||
| DILIMOT | Predicts linear motifs within protein sequences using interaction maps. | http://dilimot.russelllab.org/ | ||
| ELM Prediction | Predicts functional sites on proteins using linear motif analyses and extensive filters. | http://elm.eu.org/search.html | ||
| PAPA | Predicts prion-forming propensity for proteins by focusing on amino acid compositions. | https://combi.cs.colostate.edu/supplements/papa/ | ||
| DisoRDPbind | Predicts residues in IDRs that bind to protein, RNA, or DNA using a multilayered model. | http://biomine.cs.vcu.edu/servers/DisoRDPbind/ | ||
| LARKSdb | Predicts LARKS formation and regions involved with reversible amyloid formation and LLPS. | https://srv.mbi.ucla.edu/LARKSdb/ | ||
| MobiDB-lite | Predicts IDRs after executing 9 different predictors and filtering results for accuracy. | https://github.com/BioComputingUP/MobiDB-lite | ||
| PrionScan | Predicts prion domains on proteins using a model based on amino acid propensities. | http://webapps.bifi.es/prionscan | ||
| ProteinLIPS | Predicts locally unstable regions of proteins by identifying light interfaces of high polarity. | http://webapps.bifi.es/lips | ||
| ANCHOR | Predicts protein-binding regions within disordered proteins using pairwise energy estimation. | http://anchor.elte.hu/ | ||
| Computational tools for analyzing molecular features related to LLPS . | ||||
|---|---|---|---|---|
| Tool . | Focus and methodology . | Availability . | ||
| PLAAC | Identifies prion-like domains in proteins using a hidden Markov model. | http://plaac.wi.mit.edu/ | ||
| IUPred3 | Identifies disordered protein regions based on an energy estimation method. | https://iupred3.elte.hu/ | ||
| PSPer | First in silico tool to identify prion-like RNA-binding phase separation proteins. | https://www.bio2byte.be/b2btools/psp/ | ||
| PSPredictor | Predicts phase separation proteins using machine learning with LLPSDB. | http://www.pkumdl.cn:8000/PSPredictor/ | ||
| PScore | Predicts phase separation proteins based on pi-interaction frequencies. | https://elifesciences.org/articles/31486 | ||
| catGRANULE | Predicts granule propensity based on multiple features like RNA binding. | http://service.tartaglialab.com/update_submission/475471/abfa5da35f | ||
| dSCOPE | Predicts key regions of proteins for LLPS using a machine learning algorithm, random forest. | http://dscope.omicsbio.info/ | ||
| BIAPSS | Provides interactive statistical analyses of LLPS proteins listed in PhaSePro and LLPSDB for inferring phase-separating affinities and determining biophysical statistics of LLPS proteins. | https://biapss.chem.iastate.edu/ | ||
| PrionW | Predicts prion-like domains and amyloid nucleating cores from protein sequences. | http://bioinf.uab.cat/prionw/ | ||
| MusiteDeep | Provides a deep-learning framework for predicting and visualizing PTMs in proteins. | https://www.musite.net/ | ||
| SLiMAN | Provides a webserver for analyzing and predicting linear motifs and interactions in proteins. | http://sliman.cbs.cnrs.fr/LIMIP/limip_index.py | ||
| ESpritz | Predicts protein disorder using bidirectional recursive neural networks. | http://old.protein.bio.unipd.it/espritz/ | ||
| PLATINUM | Analyzes hydrophobic and hydrophilic characteristics of proteins and other molecules. | https://model.nmr.ru/platinum/ | ||
| PSAP | Provides a machine learning classifier that analyzes proteins and predicts phase separation. | https://github.com/Guido497/phase-separation | ||
| CIDER | Predicts and annotates protein sequences for IDRs/IDPs for their conformational properties. | http://pappulab.wustl.edu/CIDER/ | ||
| DisMeta | Provides a meta-server that has predictors for disordered and low-complexity regions in proteins. | https://montelionelab.chem.rpi.edu/dismeta/ | ||
| DILIMOT | Predicts linear motifs within protein sequences using interaction maps. | http://dilimot.russelllab.org/ | ||
| ELM Prediction | Predicts functional sites on proteins using linear motif analyses and extensive filters. | http://elm.eu.org/search.html | ||
| PAPA | Predicts prion-forming propensity for proteins by focusing on amino acid compositions. | https://combi.cs.colostate.edu/supplements/papa/ | ||
| DisoRDPbind | Predicts residues in IDRs that bind to protein, RNA, or DNA using a multilayered model. | http://biomine.cs.vcu.edu/servers/DisoRDPbind/ | ||
| LARKSdb | Predicts LARKS formation and regions involved with reversible amyloid formation and LLPS. | https://srv.mbi.ucla.edu/LARKSdb/ | ||
| MobiDB-lite | Predicts IDRs after executing 9 different predictors and filtering results for accuracy. | https://github.com/BioComputingUP/MobiDB-lite | ||
| PrionScan | Predicts prion domains on proteins using a model based on amino acid propensities. | http://webapps.bifi.es/prionscan | ||
| ProteinLIPS | Predicts locally unstable regions of proteins by identifying light interfaces of high polarity. | http://webapps.bifi.es/lips | ||
| ANCHOR | Predicts protein-binding regions within disordered proteins using pairwise energy estimation. | http://anchor.elte.hu/ | ||
Abbreviations: IDP, intrinsically disordered protein; PTM, posttranslational modification.
LLPS is enriched in cancer. A, Of the 287 genes with annotated cancer hallmarks in COSMIC, 53 undergo LLPS (vertical line), significantly more than observed when selecting 287 random genes with GO annotated biological processes (histogram). B, Comparison of enrichment for cancer-associated genes from A with those linked to other human diseases as well as genes that are associated with multiple diseases.
LLPS is enriched in cancer. A, Of the 287 genes with annotated cancer hallmarks in COSMIC, 53 undergo LLPS (vertical line), significantly more than observed when selecting 287 random genes with GO annotated biological processes (histogram). B, Comparison of enrichment for cancer-associated genes from A with those linked to other human diseases as well as genes that are associated with multiple diseases.
Cancer hallmarks represented by phase separation genes. A, Enrichment of specific cancer hallmarks from LLPS genes. B, Bipartite network representing LLPS genes and corresponding pathways from A.
Cancer hallmarks represented by phase separation genes. A, Enrichment of specific cancer hallmarks from LLPS genes. B, Bipartite network representing LLPS genes and corresponding pathways from A.
DERANGEMENT OF PHASE SEPARATION IN CANCER
Spatiotemporal Reorganization of Cellular Material
Despite enrichment for phase separation among cancer genes, and the expanding appreciation for this phenomenon as an organizing principle in cell biology, only relatively recently have investigators begun to examine connections between phase separation and malignancy. The maintenance of a proper proteostatic equilibrium is essential for healthy cells (33). Oncogenic transformation places many additional demands on protein quality control systems, including molecular chaperones, the ubiquitin proteasome system, and autophagy (34, 35). Hubs of many of these protein quality control systems, including the autophagosome and proteasome, have now been proposed to be organized via the principles of phase separation (36, 37). Cancer-specific mutations in SPOP, a tumor suppressor and adaptor for the BTB–CUL3–RBX1 E3 ubiquitin ligase responsible for proteasomal targeting, provide a striking example. Mutations in SPOP are associated with many types of cancer (e.g., prostate, breast, endometrial, gastric, kidney, and ovarian). It has long been known that these mutations can impair substrate recruitment by the ligase. But deeper analysis has recently revealed that substrate interactions with the SPOP adapter drive phase separation that is critical for E3 ligase activity and ensuing proteolytic turnover (38). Inhibition of these interactions, as occurs for SPOP mutations associated with diverse cancers, thus leads to the accumulation of proto-oncogenic proteins (38).
In addition to its role in substrate-directed phase separation, SPOP regulates the assembly of stress granules, which form via phase separation (8). Stress granules play key roles in cellular adaptation to stress and are often upregulated in cancers. Analysis of SPOP-interacting proteins provided a clue to a possible mechanism. The SPOP interactor Caprin1 promotes nucleation of stress granules and is upregulated in prostate cancer (where SPOP is most frequently mutated; ref. 39). This arises because mutant SPOP can no longer recognize Caprin1 as a substrate, leading to its accumulation and subsequent increases in stress granule assembly. As a collateral consequence, cancer cells gain the ability to adapt and even thrive under stressful environmental conditions.
Cancer Fusions
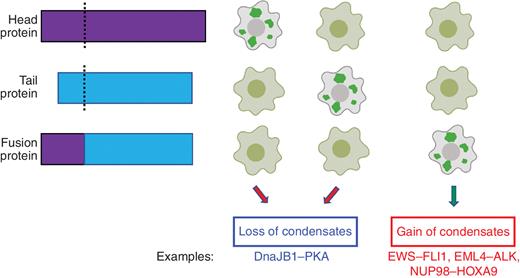
In addition to aberrations in phase separation induced by mutations to single genes, cancer fusions can exert a strong impact on this emergent property, leading to either gain or loss of condensates (Fig. 4). Cancer-related fusion proteins often rewire protein interaction networks (40–43). Well-known examples that alter phase separation include members of the FET protein family (EWS and FUS) and the RTK family (ALK and RET). Fusions of EWS and FUS to transcription factors (TF) are known drivers of both Ewing sarcoma and myxoid liposarcomas (44, 45). Aberrant phase separation driven by the prion-like domain within the disordered sequence of EWS–FLI1 promotes altered BAF-dependent chromatin remodeling and the establishment of oncogenic gene expression programs (44). The formation of phase-separated foci of EML4–ALK preserves the kinase activity of ALK but excludes repressive factors, mediating oncogenic signaling involving MAPK, PLCγ, and PI3K pathways (46). These oncogenic fusion condensates locally concentrate the RAS-activating complex and activate the MAPK signaling cascade via membraneless protein granules (47). In addition, NUP98 fusion oncoproteins such as NUP98–HOXA9, which is associated with pediatric leukemias, can form nuclear puncta through combined homotypic and heterotypic interactions, fueling aberrant transcriptional rewiring and oncogenic transformation (41–43). Finally, the oncogenic fusion DnaJB1–PKA, which is frequently detected in liver cancer, abolishes PKA-mediated phase separation that is critical for cAMP compartmentation and cancer signaling (48).
Alteration of phase separation behavior by fusion proteins. Fusion proteins (bottom) formed by aberrant joining of head protein (top) and tail protein (middle) at their breakpoints (dash lines). The corresponding phase separation phenotype (loss or gain of condensates) for each of these proteins is shown on the right. Each column of cell images represents one group for comparative purpose (cells harboring fusion proteins or their corresponding head or tail parent proteins). Specific examples for the gain or loss of condensates from the literature are also shown.
Alteration of phase separation behavior by fusion proteins. Fusion proteins (bottom) formed by aberrant joining of head protein (top) and tail protein (middle) at their breakpoints (dash lines). The corresponding phase separation phenotype (loss or gain of condensates) for each of these proteins is shown on the right. Each column of cell images represents one group for comparative purpose (cells harboring fusion proteins or their corresponding head or tail parent proteins). Specific examples for the gain or loss of condensates from the literature are also shown.
Cancer-related fusion proteins are often enriched in IDRs. Dysregulation of IDRs could cause errors in the location and trafficking of molecules in the cell (40, 49). There has been a rapidly increasing appreciation of the role of IDRs—long considered the dark matter of the proteome—in the proteome (40). IDRs evolve more rapidly than ordered protein regions (23). This has primarily been attributed to reduced constraints on preserving globular structures, but others have argued that IDRs can frequently drive the adaptive evolution of cells that harbor them (50). Fusion of IDRs can occur via chromosomal translocations that often drive cancers; the relatively high rates at which such fusion events occur and their potential to rewire intracellular organization, signaling, and gene expression may provide a crucible for rapid cancer clonal evolution (51).
PHASE SEPARATION AND AGGREGATION IN CANCER-RELATED PROCESSES
DNA Damage Response
Ionizing radiation and radiomimetic substances such as DNA cross-linkers and alkylating agents cause double-strand breaks (DSB) in DNA. If left unrepaired, DSBs can drive the genomic instability that is a hallmark of most tumors. The rapid, site-specific, and transient assembly of DNA repair factors at these sites of damage is essential to repair DSBs and prevent genomic instability. A multivalent, polyanionic, nucleic acid–like molecule called poly-ADP ribose (PAR) is synthesized at sites of DNA damage and plays a crucial role in the DNA damage signaling network (52). Because multivalency in biomolecules often fuels phase separation, the capacity of PAR to influence such spatiotemporal reorganization was investigated. Live cell imaging demonstrated that PAR nucleates the phase separation of several intrinsically disordered proteins at sites of DNA damage (53). Indeed, this is one of the earliest steps in DNA damage signaling. Notable among these proteins are FET family members such as FUS, EWS, and TAF15. These proteins harbor repetitive RGG motifs that are cationic at cellular pH, favoring interaction with negatively charged PAR, which drives phase separation. Importantly, these liquid droplets concentrate a selective cohort of proteins and do not readily mix with foci formed at DNA damage sites by the genomic caretaker protein 53BP1 (p53-binding protein 1; refs. 53, 54).
53BP1 is a master regulator of DSB signaling and repair by nonhomologous end joining (NHEJ). During the G1 phase of the cell cycle, in the absence of a sister chromatid, NHEJ pathways are the primary mode of DNA repair responsible for placing the broken DNA ends in proximity and ligating them. Spatiotemporally distinct from PAR-driven phase separation events, 53BP1 also undergoes DNA damage–induced LLPS. This features a hallmark of liquid droplets: frequent fusions and occasional fission of these foci. Condensation is primarily driven by the C-terminal oligomerization domain of 53BP1. Disruption of 53BP1 condensates destabilizes interactions with p53, reducing its ability to induce expression of downstream effectors such as p21 (55). 53BP1 condensates have therefore been implicated in coordinating local DNA damage recognition with global changes in downstream gene expression programs (55). A critical role of a noncoding damage-inducible long noncoding RNA transcribed at DNA damage sites in controlling the liquid-like properties of 53BP1 condensates has also been investigated (56). These few examples already suggest that phase separation plays a critical role in the DNA damage response. Indeed, proteins involved in DNA repair often form dynamic foci associated with activity (57, 58). Further investigations will no doubt continue to shed light on the role of posttranslational modifications in regulating such phenomena and how this process is altered in, or perhaps even acts as a cause of, oncogenic transformation.
Classic Proliferation Marker Ki-67
Histologic markers of cancer were the gold standard in the clinic prior to the advent of genetic biomarkers and remain widely used. For example, the Ki-67 labeling index formulated on the eponymous protein is a classic marker for proliferation (59, 60). It has been routinely used in the clinic to identify and stage a wide variety of cancers ranging from breast cancers to neuroendocrine tumors (61, 62). Despite its enormous importance, little was known about the cellular mechanism of the Ki-67 protein. Recently, in a targeted siRNA screen, Ki-67 was found to play a key role in maintaining chromosome separation (63), consistent with its localization to the chromosome periphery (64). Further investigation revealed that Ki-67 was required to maintain spatial separation after nuclear envelope breakdown. The Ki-67 protein consists of an N-terminal region comprising a phosphopeptide-binding Forkhead-associated domain and a protein phosphatase 1–binding site, a central region comprising 16 tandem repeats, and a C-terminal region enriched in leucine–arginine pairs (65). The C-terminal region interacts with chromatin and is essential for the individualization of mitotic chromosomes. However, it is not sufficient on its own. Mutational analyses revealed that coupling the chromatin-targeting region with either the N-terminal region or the central region with tandem repeats restored this function. Strikingly, even a scrambled N-terminal region coupled to the essential chromatin-targeting region is sufficient for restoring its function. This leads to the model in which the general physicochemical properties of these regions and not their sequence are of primary importance in its function. The existing model suggests that Ki-67 behaves as an amphiphilic surfactant, coating the chromosome and forming a phase boundary that allows appropriate spatial separation of chromosomes (63). This example highlights the unexpected ways in which the emergent properties of phase separation lie at the heart of many hallmarks of cancer.
p53
The ability of MLOs to compartmentalize cellular space occurs in the cytoplasm as well as the nucleoplasm. Cajal bodies and promyelocytic leukemia (PML) nuclear bodies are examples of LLPS phenomena in the nucleoplasm. The PML protein provides the primary scaffold for these micron-sized, subnuclear organelles, and additional multivalent interactions further stabilize them (66). Several lines of evidence implicate PML nuclear bodies as key mediators of the tumor suppressor functions of p53. Transcriptional activation of classic p53 target genes such as Bax and p21 is severely impaired when p53 does not localize to PML bodies (67). Furthermore, PML−/− cells have a reduced propensity to senesce or undergo apoptosis, even upon p53 activation (68). In the MCF-7 breast carcinoma cell line, the p53 signaling cascade is impaired by PML knockdown (69). The PML protein also protects p53 from proteasomal degradation. Under conditions of DNA damage stress, PML sequesters Mdm2 in the nucleolus, thus preventing p53 ubiquitination and degradation (70). WRAP53, a gene implicated in cancer development, encodes both an antisense transcript to p53 and a WD40 repeat containing protein that promotes the formation of Cajal bodies (71). Although there are obvious implications of this antisense transcript regulating p53 activity, the connection to phase-separated Cajal bodies in this process remains to be investigated in depth.
In addition to functionally associating with phase-separated biomolecular condensates, wild-type and certain p53 mutants have been demonstrated to undergo liquid-like phase separation on their own, in a protein-autonomous manner (72, 73). In vitro experiments suggest that the liquid-like properties of p53 can be modulated by interaction with ATP and nucleic acids, and a phosphomimetic mutant of the protein p53S392E abrogates droplet formation (72). A few other mutations that commonly occur in patients have been found to render the protein amyloidogenic (74). The amyloid fibers formed by the mutant protein can sequester wild-type p53, leading to a dominant-negative effect and loss of p53 function (75). Peptide inhibitors of amyloid assembly block proliferation and restore p53 tumor suppression in ovarian carcinoma cells harboring these mutations (76).
EMERGENT ONCOGENIC PROPERTIES OF PROTEIN–NUCLEIC ACID CONDENSATES
Regulation of Gene Expression
Dysregulation of gene expression is a pervasive theme across all cancers. A string of recent articles have begun to delineate a potential role for biomolecular condensates in transcriptional gene control. Comparative analyses between healthy and cancerous cells have established that gene deserts around oncogenes such as MYC acquire many clusters of enhancer elements known as superenhancers (77). Recent findings have suggested that superenhancers and many of the integral components of the transcription machinery that interact with them including Mediator complex proteins, Brd4, as well as RNA Pol II can undergo phase separation and form MLOs (78–80), although there has been vigorous debate in the literature about whether some of these structures should instead be defined as “hubs” (81).
Phase separation has been speculated to explain many features of transcriptional control including local concentration of many of the factors necessary for gene activation. IDRs in these proteins are thought to be the driving force for the formation of these condensates. In this context, DNA translocations that are common in cancer are often associated with the acquisition of superenhancers at driver oncogenes (77). Furthermore, the presence of such IDRs on many TFs across evolution and their frequent shuffling in cancer fusions collectively suggest an important role for IDR-driven phase separation in cancer (82).
Another series of investigations in the model eukaryote Saccharomyces cerevisiae have revealed how some intrinsically disordered TFs and RNA-binding proteins (RBP) form gel-like assemblies that can propagate in a prion-like fashion, acting as protein-based epigenetic elements that can transform gene expression over medium to long biological timescales. Even transient overexpression of these TFs and RBPs, as can occur for their human orthologs in cancer, can trigger the formation of self-templating protein assemblies that rewire the gene expression landscape for many cell divisions (24, 83, 84). Studies with the RBP Vts1/Smaug, whose human ortholog SAMD4A is overexpressed in chronic lymphocytic leukemia, revealed that its transient overexpression drove the formation of condensates that hyperactivate the protein's ability to degrade target RNAs (24). Consequently, cells alter how they couple metabolic information to decisions about proliferation and differentiation (85). Notably, SAMD4A retains the capacity to form condensates with heritable prion-like properties (24). Likewise, the Snt1 subunit of the Set3 histone deacetylase complex (ortholog of the NCOR–SMRT deacetylase complex that is dysregulated in many endocrine-related cancers; ref. 86) can self-assemble into self-templating gels, driving epigenetic inheritance of active chromatin and robust drug resistance (83). These findings have provoked efforts from us and others to understand how condensation of gene regulators can affect phenotype over medium to long biological timescales.
Alteration of Environmental Sensing and Response
Biochemical selectivity, a foundational requirement for the precise regulation of signaling pathways, can also be afforded by phase-separated condensates (87). This may have important implications for cancer, as the rewiring of metabolic equilibria and biomolecular interactions play a key role in driving the initiation and progression of the disease. In line with this, phase separation of T-cell receptor signaling protein components fuels the formation of biochemically selective compartments (i.e., including kinases while excluding phosphatases) that play key roles across many cancers (88). Thus, alterations in phase separation can drive changes in biochemical selectivity relevant to cancer signaling.
Many signaling pathways rely on secondary messengers to relay and amplify responses across the cell. As one example of how phase separation can affect this behavior, DNA-induced condensation can promote the production of secondary messengers in cGAMP synthase (cGAS) signaling (89). In addition to secondary messenger production, phase separation plays important roles in target gene activation. For example, activation of the HIPPO pathway, which is critical for cell growth and proliferation, is mediated by phase separation of pathway-specific transcription coactivators (90, 91). Because cancer is often driven by changes in gene expression, it is tempting to speculate that altered phase separation of key TFs and their regulators might provide a common mechanism through which this can be achieved.
Although the aberrant activation of signaling pathways is often observed in cancer, it is common that cancer cells lose the capacity to turn off signaling pathways via negative feedback systems. Here, too, phase separation can play an important regulatory role. For example, the WNT/beta-catenin pathway is negatively regulated through the beta-catenin destruction complex. Recent work has shown that the destruction complex, which promotes the proteolysis of beta-catenin to keep its levels low in the absence of WNT, forms via phase separation (92). Furthermore, cytoplasmic DACT1 condensates that are induced by TGFβ repress WNT signaling. These DACT1 condensates are maintained in vivo and are associated with the bone metastasis of breast and prostate cancers (93). Future work will be needed to fully comprehend how phase separation functions in the negative regulation of signaling pathways in both normal biology and disease states.
Alteration of Cellular Metabolism
One of the classic examples of alteration of metabolic flux in cancer cells was described by Otto Warburg and colleagues. The harnessing of glycolytic pathways over oxidative phosphorylation by cancer cells has been extensively described (94). Recent experiments in model organisms such as yeast, Caenorhabditis elegans, and mammalian cells have demonstrated that many of the enzymes involved in glycolysis assemble into membraneless cytoplasmic organelles, called glycolytic bodies (G-bodies), under hypoxic stress (95, 96). In yeast, G-bodies form when oxidative phosphorylation is inhibited, and this improves their fitness under hypoxia (97). G-bodies arise under hypoxic conditions in HepG2 cells, a hepatocellular carcinoma cell line (96). These subcellular organelles accelerate glucose consumption and enhance glycolytic flux by increasing the local concentration of the glycolysis enzymes. The enzyme phosphofructokinase, Pfk2 in yeast and PFKL in humans, drives the formation of G-bodies (96). The N-terminal disordered region of this protein is important for their formation (97). Recently, unbiased searches for RNA-binding activity have demonstrated that many metabolic enzymes moonlight as RBPs. In fact, many RNAs are strongly associated with G-bodies as well and contribute to their structural integrity; targeting RNase to G-bodies disrupts them (97). From a structural standpoint, G-bodies do not resemble liquids. Rather, they are a gel-like species emphasizing the diverse palette of material properties that can contribute to function in the complex cellular environment. This example may be just the tip of the iceberg. Phase separation of proteins (98, 99) associated with oxidative pathways can sense cellular redox states, and some have been demonstrated to alter the flux of key metabolic regulators such as the TORC1 complex (100, 101).
Alternative Splicing
The modular addition of IDRs to protein sequences via alternative splicing offers an intriguing path to diversify protein function. Given the ability of IDRs to endow proteins with the ability to phase separate, this leads to a natural diversification of the material properties of condensates formed by the protein. This is perhaps best exemplified by heterogeneous nuclear ribonucleoprotein (hnRNP) A and D family members (102). These proteins play diverse roles in mammalian RNA metabolism, and hnRNPDL in particular is upregulated in a variety of cancers including prostate, CML, hepatocellular carcinoma, and colon. hnRNPDL has three isoforms: DL1, DL2, and DL3. These isoforms differ in the way the IDRs are arranged with consequent changes in their cellular behavior: DL3 does not harbor any IDRs and is completely soluble, DL2 harbors the C-terminal IDR, and DL1 harbors IDRs at both termini (103). The C-terminal IDR is tyrosine rich and confers amyloidogenic properties to the DL2 isoform. Strikingly, the presence of the arginine-rich N-terminal IDR in the DL1 isoform abrogates fibril formation and enhances the formation of large amorphous condensates. It has been suggested that the structural differences among the isoforms arise from the differences between pi–pi interactions among aromatic residues in the dominant DL2 isoform and pi–cation interactions in DL1. The structural differences also correlate with differential localization within the cell. Certain point mutations in the C-terminal IDR increase the aggregation propensity of both DL1 and DL2 isoforms and are associated with an autosomal dominant form of muscular dystrophy. Interestingly, SRSF, which is a key upstream regulator of alternative splicing of hnRNPDL, is also dysregulated in multiple cancers (103, 104).
TARGETING PHASE SEPARATION AS A THERAPEUTIC STRATEGY
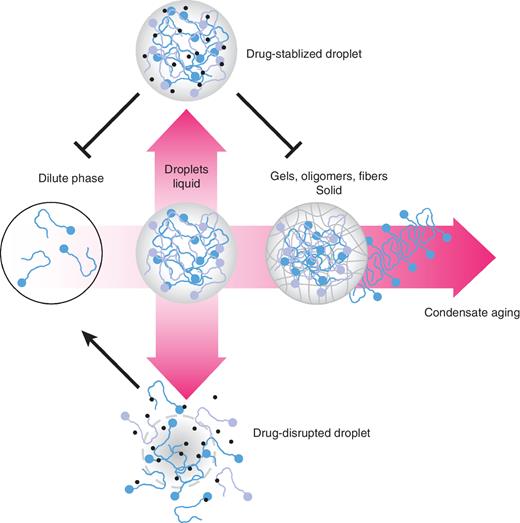
Phase separation provides several new avenues for potential therapeutic intervention. Despite the widespread occurrence of higher order protein aggregates in many diseases of aging, therapeutic targeting of the associative properties of biomolecules has been notoriously difficult, with the exception of groundbreaking approaches to develop chemical chaperones/stabilizers for specific misfolded proteins (105, 106). Therapeutic targeting of phase separation could significantly expand the palette of drug development strategies for TFs and other proteins involved in gene regulation that have been notoriously difficult to drug. Yet doing so will require a deepened consideration of physicochemical principles that go beyond the lock and key approaches that are often deployed to target ordered proteins. One strategy for direct targeting of phase-separated entities relies on the differential partitioning of a small molecule or a biologic between the dilute and dense phases (Fig. 5). The disruption of many LLPS entities by 1,6-hexanediol first shed light on this possibility. From a therapeutic standpoint, recent work has demonstrated the feasibility of several compounds to disrupt the RNA-dependent recruitment of several RBPs—namely, TDP-43, FUS, and hnRNPA2B1—into stress granules (107). Furthermore, drugs such as cisplatin can be enriched within condensates (108). A recent chemical screen has also identified small molecules that can modulate the phase separation properties of p53 mutants (109), foreshadowing a new genre of therapeutics for future cancer treatment. However, many challenges remain to overcome, including the development of molecules that can selectively target different phase-separated condensates in the same cellular environment. Furthermore, the differential sequestration of drugs within condensates raises the possibility that resistance may occur through mutations that alter phase separation.
Therapies targeting condensates. The prevalence of cancer genes that form condensates presents an opportunity to develop new classes of therapeutics that modulate the phase separation of their targets. New drugs might be made to disrupt or prevent specific molecules from forming liquid droplets (bottom arrow), while others might stabilize the liquid droplet, blocking its dissolution or maturation into solid-phase MLOs.
Therapies targeting condensates. The prevalence of cancer genes that form condensates presents an opportunity to develop new classes of therapeutics that modulate the phase separation of their targets. New drugs might be made to disrupt or prevent specific molecules from forming liquid droplets (bottom arrow), while others might stabilize the liquid droplet, blocking its dissolution or maturation into solid-phase MLOs.
The functional role of mutations in altering phase separation needs to be systematically evaluated, especially in cancer.
Cancer gene fusions could have a profound impact on phase separation and indeed use this property to drive cancer initiation and progression. Systematic efforts are needed to understand this mechanism.
Cancer mutations could drive either gain or loss of condensates, contributing to disease.
Many missense mutations could affect phase separation, through rewiring relevant signaling pathways, and known disease mutations may function by altering the presence or composition of condensates.
The extent to which protein folding and stability contribute to phase separation activities is mostly unknown.
Most importantly, many disease alleles, especially cancer mutants, could be dominant, sequestering wild-type counterparts to form condensates. This remarkable behavior needs to be systematically examined, which could reveal fundamental principles of genetic inheritance.
Authors’ Disclosures
W.J. Zheng reports grants from the NIH and the Cancer Prevention & Research Institute of Texas during the conduct of the study. G.B. Mills is on scientific advisory boards for Amphista, AstraZeneca, Chrysalis Biotechnology, GSK, ImmunoMET, Ionis, Lilly, PDX Pharmaceuticals, Signalchem Lifesciences, Symphogen, Tarveda, Turbine, and Zentalis Pharmaceuticals, and reports other support from Catena, Genentech, NanoString Center of Excellence, and Ionis (provision of tool compounds) during the conduct of the study, as well as a patent for an HRD assay to Myriad Genetics and a patent for DSP to NanoString. D.F. Jarosz reports grants from the NIH, the David and Lucile Packard Foundation, and the Vallee Foundation during the conduct of the study, as well as personal fees from Transition Bio outside the submitted work. No disclosures were reported by the other authors.
Acknowledgments
This work was supported by Komen Foundation grant CCR19609287 (to S.S. Yi); National Institute of General Medical Sciences grants R35GM133658 (to S.S. Yi) and R35GM137836 (to N. Sahni); NIH grant DP2-GM119140 (to D.F. Jarosz); Department of Defense fund W81XWH-18-PRCRP-CDA CA181455 (to N. Sahni); and NCI grants P50CA217685 and UO1CA217842 (to G.B. Mills). N. Sahni is a CPRIT Scholar in Cancer Research with funding from the Cancer Prevention & Research Institute of Texas (CPRIT) New Investigator Grant RR160021 and Research Grant RP220292. S.S. Yi was also supported by the Scialog program sponsored jointly by the Research Corporation for Science Advancement and the Gordon and Betty Moore Foundation (award number 28418) as well as a pilot project program sponsored jointly by the Oden Institute, MD Anderson, and Texas Advanced Computing Center. G.B. Mills also received a kind gift from the Miriam and Sheldon Adelson Medical Research Foundation and the Breast Cancer Research Foundation. D.J. McGrail was supported by NCI grant K99CA240689. A.K. Chakravarty was supported as a Howard Hughes Medical Institute fellow of the Damon Runyon Cancer Research Foundation (DRG2221-15) and by an NIH Pathway to Independence Award (1K99GM128180-01). T.M. Lozanoski was supported by a National Science Foundation (NSF) graduate research fellowship. D.F. Jarosz was also supported as a Searle Scholar, as a Kimmel Scholar, as a Vallee Scholar, by a Science and Engineering Fellowship from the David and Lucile Packard Foundation, and by an NSF CAREER Award (1453762) from the NSF. W.J. Zheng and D.J.H. Shih were partly supported by the NIH through grant 1UL1TR003167 and CPRIT through grant RP170668. We also acknowledge support from BioRender in figure generation.