Abstract
Epithelial-to-mesenchymal transition (EMT) renders epithelial cells migratory properties. While epigenetic and splicing changes have been implicated in EMT, the mechanisms governing their crosstalk remain poorly understood. Here we discovered that a C2H2 zinc finger protein, ZNF827, is strongly induced during various contexts of EMT, including in brain development and breast cancer metastasis, and is required for the molecular and phenotypic changes underlying EMT in these processes. Mechanistically, ZNF827 mediated these responses by orchestrating a large-scale remodelling of the splicing landscape by recruiting HDAC1 for epigenetic modulation of distinct genomic loci, thereby slowing RNA polymerase II progression and altering the splicing of genes encoding key EMT regulators in cis. Our findings reveal an unprecedented complexity of crosstalk between epigenetic landscape and splicing programme in governing EMT and identify ZNF827 as a master regulator coupling these processes during EMT in brain development and breast cancer metastasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The sequencing data generated in this study have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession code of GSE173874. MS data have been deposited in PeptideAtlas (http://www.peptideatlas.org/) with the primary accession code PASS01764. Previously published datasets were re-analysed here, including TCGA Research Network (https://www.cancer.gov/tcga), breast cancer (BRCA-TCGA) transcriptome data. Any other data supporting the findings of this study are available upon request. Source data are provided with this paper.
References
Yang, J. et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020).
Kalluri, R. & Weinberg, R. A. The basics of epithelial–mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009).
Burger, G. A., Danen, E. H. J. & Beltman, J. B. Deciphering epithelial–mesenchymal transition regulatory networks in cancer through computational approaches. Front. Oncol. 7, 162–162 (2017).
De Craene, B. & Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 13, 97–110 (2013).
Braunschweig, U., Gueroussov, S., Plocik, A. M., Graveley, B. R. & Blencowe, B. J. Dynamic integration of splicing within gene regulatory pathways. Cell 152, 1252–1269 (2013).
Warzecha, C. C. et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 29, 3286–3300 (2010).
Shapiro, I. M. et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 7, e1002218 (2011).
Pillman, K. A. et al. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 37, e99016 (2018).
Luco, R. F., Allo, M., Schor, I. E., Kornblihtt, A. R. & Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 144, 16–26 (2011).
Brody, Y. et al. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 9, e1000573 (2011).
Saldi, T., Cortazar, M. A., Sheridan, R. M. & Bentley, D. L. Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol. 428, 2623–2635 (2016).
Schmitges, F. W. et al. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 26, 1742–1752 (2016).
Conomos, D., Reddel, R. R. & Pickett, H. A. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 21, 760–770 (2014).
Yang, S. F., Sun, A. A., Shi, Y., Li, F. & Pickett, H. A. Structural and functional characterization of the RBBP4–ZNF827 interaction and its role in NuRD recruitment to telomeres. Biochem. J. 475, 2667–2679 (2018).
Sahu, S. K. et al. JNK-dependent gene regulatory circuitry governs mesenchymal fate. EMBO J. 34, 2162–2181 (2015).
Janda, E. et al. Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J. Cell Biol. 156, 299–313 (2002).
Iorns, E. et al. A new mouse model for the study of human breast cancer metastasis. PLoS ONE 7, e47995 (2012).
Tang, Z. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017).
Yan, Z. et al. OSbrca: a web server for breast cancer prognostic biomarker investigation with massive data from tens of cohorts. Front. Oncol. 9, 1349 (2019).
Camargo Ortega, G. et al. The centrosome protein AKNA regulates neurogenesis via microtubule organization. Nature 567, 113–117 (2019).
Bibel, M., Richter, J., Lacroix, E. & Barde, Y. A. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc. 2, 1034–1043 (2007).
Telley, L. et al. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351, 1443–1446 (2016).
Saito, T. In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 1, 1552–1558 (2006).
Najafabadi, H. S. et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33, 555–562 (2015).
Mani, S. A. et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Bulyk, M. L., Huang, X., Choo, Y. & Church, G. M. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl Acad. Sci. USA 98, 7158–7163 (2001).
Persikov, A. V. et al. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 43, 1965–1984 (2015).
Xia, B. et al. NR2F2 plays a major role in insulin-induced epithelial–mesenchymal transition in breast cancer cells. BMC Cancer 20, 626 (2020).
Han, H. et al. Multilayered control of alternative splicing regulatory networks by transcription factors. Mol. Cell 65, 539–553 e537 (2017).
Nieto, M. A., Huang, R. Y., Jackson, R. A. & Thiery, J. P. Emt: 2016. Cell 166, 21–45 (2016).
Li, J. et al. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. eLife 7, e37184 (2018).
Yang, Y. et al. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol. Cell Biol. 36, 1704–1719 (2016).
Conn, S. J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015).
Fong, N. et al. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 28, 2663–2676 (2014).
Barbieri, E. et al. Rapid and scalable profiling of nascent RNA with fastGRO. Cell Rep. 33, 108373 (2020).
Singh, J. & Padgett, R. A. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 16, 1128–1133 (2009).
Mrkvicova, A. et al. The effect of sodium butyrate and cisplatin on expression of EMT markers. PLoS ONE 14, e0210889 (2019).
Li, L. et al. Targeting the ERG oncogene with splice-switching oligonucleotides as a novel therapeutic strategy in prostate cancer. Br. J. Cancer 123, 1024–1032 (2020).
Havens, M. A. & Hastings, M. L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 44, 6549–6563 (2016).
Warns, J. A., Davie, J. R. & Dhasarathy, A. Connecting the dots: chromatin and alternative splicing in EMT. Biochem. Cell Biol. 94, 12–25 (2015).
Wawruszak, A. et al. Histone deacetylase inhibitors and phenotypical transformation of cancer cells. Cancers 11, 148 (2019).
Hu, Y., Nie, Q., Dai, M., Chen, F. & Wu, H. Histone deacetylases inhibit the snail2-mediated EMT during metastasis of hepatocellular carcinoma cells. Front. Cell Dev. Biol. 8, 752 (2020).
Shu, P. et al. MicroRNA-214 modulates neural progenitor cell differentiation by targeting Quaking during cerebral cortex development. Sci. Rep. 7, 8014 (2017).
Sahu, S. K. et al. FBXO32 promotes microenvironment underlying epithelial-mesenchymal transition via CtBP1 during tumour metastasis and brain development. Nat. Commun. 8, 1523 (2017).
Hollensen, A. K. et al. circZNF827 nucleates a transcription inhibitory complex to balance neuronal differentiation. eLife 9, e58478 (2020).
Cerami, E. et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Lou, Y. et al. Epithelial–mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin. Cancer Res. 22, 3630–3642 (2016).
Kim, H. K. et al. Deep learning improves prediction of CRISPR–Cpf1 guide RNA activity. Nat. Biotechnol. 36, 239–241 (2018).
Tiwari, N. et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23, 768–783 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Kaimal, V., Bardes, E. E., Tabar, S. C., Jegga, A. G. & Aronow, B. J. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 38, W96–W102 (2010).
Sherman, B.T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
Guberman, J. M. et al. BioMart Central Portal: an open database network for the biological community. Database 2011, bar041 (2011).
Yates, A. et al. Ensembl 2016. Nucleic Acids Res. 44, D710–D716 (2016).
Ong, S. E. & Mann, M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 (2006).
Michalski, A. et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell Proteomics 10, M111 011015 (2011).
Acknowledgements
We thank the members of the Tiwari lab for their cooperation and critical feedback throughout this study. We thank N. Hah from Salk Institute for Biological Studies for her feedback on fastGRO. The support from the Facilities of the Queen’s University Belfast is gratefully acknowledged. This study was supported by the Deutsche Forschungsgemeinschaft TI 799/1-3, Wilhelm Sander Stiftung 2012.009.1 and 2012.009.2 and ICURe to V.K.T. and the Marie Curie (PCIG12-GA-2012-334000) and ANR programme (CT-141033_ANR-16-CE12-0012-01) to R.F.L.
Author information
Authors and Affiliations
Contributions
S.K.S. designed and performed experiments, analysed data and wrote the manuscript. E.A. and M.I. performed computational analysis. A.M. and D.L. performed experiments and analysed data. N.T. provided conceptual input and wrote the manuscript. A.S. provided input to the brain development study. S.S. and M.D. provided help with in vivo tumourigenicity assays and imaging. R.L. provided critical input regarding alternative splicing analysis. J.C.I.B. provided support and reagents for this study. V.T. designed the study, analysed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Muh-Hwa Yang, Philip Gregory and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Model systems for EMT.
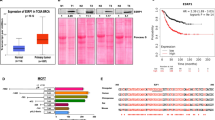
(a) Heatmap showing expression of EMT markers in TCGA samples. The EMT status of samples are calculated based on the EMT-score and highlighted as EMT-Low (Epithelial) (n = 578), Pre-EMT (early mesenchymal) (n = 221) and EMT-high (mesenchymal) (n = 301) groups. The samples highlighted with green and brown annotations are top 25 % of each EMT-Low and EMT-high category. (b) Spearman correlation plot constructed using Z–scores values showing correlation among EMT markers. The Significance Test was performed for each pair of markers with a 95% confidence level and P-value <0.05. The boxes with orange color depict positive correlation and blue boxes depict negative correlation. (c) ZNF827 mRNA levels were measured by RT-PCR in two human breast cancer epithelial cell lines (MCF7 and MDA-MB-361) and two mesenchymal cell lines (MDA-MB-231 and BT547). Error bars represent the mean ± SEM of three independent biological replicates. (d) RNA-seq data showing mRNA level of key EMT markers in NMuMG cells undergoing EMT, the y-axis represents the normalized tag count. Error bars represent the mean ± SEM of three independent biological replicates. (e) Representative bright-field images of NMuMG cells undergoing TGF-β induced EMT. (f) RNA-seq data showing mRNA level of key EMT markers in HMLE cells undergoing EMT, the y-axis represents the normalized tag count. Error bars represent the mean ± SEM of three independent biological replicates. (g) Representative bright-field images of HMLE cells undergoing TGF-β induced EMT. e and g are representative of three independent biological replicates. Source numerical data are provided in Source Data Ext. Figure 1.
Extended Data Fig. 2 ZNF827 is induced during EMT.
(a) RNA-seq data showing mRNA levels of Zfp827 in NMuMG cells undergoing TGF-β-induced EMT, the y-axis represents the normalized tag count. (b-c) RT-PCR measurements of Zfp827/ ZNF827 mRNA levels in EpRas (b), and HMEC (c) cells during TGF-β-induced EMT. RNA levels are shown relative to Ctcf/ CTCF mRNA levels. (d) mRNA levels of Zfp827 were measured relative to Ctcf using RT-PCR in NMuMG cells undergoing TGF-β-induced EMT and treated with inhibitors of various Map Kinases or depleted for Smad4. (e-f) Representative immuno-fluorescence and bright-field images of the localization and expression levels of EMT marker proteins in NMuMG (e) and human primary mammary epithelial (HMEC) (f) cells which are either untreated or undergoing TGF-β-induced EMT and treated with either non-targeting control (siControl) or siRNA against ZNF827 (siZNF827) with pools of four siRNAs. Staining was performed to assess the expression with antibodies against the epithelial markers E-cadherin and ZO1, the mesenchymal markers Fibronectin and Phalloidin (to visualize the actin cytoskeleton) and Paxillin (to detect focal adhesion plaques). Error bars represent the SEM of three independent biological replicates. Data represent three biological replicates. (g) A representative immunoblot of two independent biological replicates showed knockdown efficiency of ZNF827 in HMEC cells undergoing TGF-β-induced EMT at day 4 (d4). LMNB was used as a loading control. Error bars represent the mean ± SEM of three independent biological replicates for a-c. Statistical analyses were performed using a two-tailed unpaired Student’s t-test *p < 0.05. n= two independent biological replicates for d. Source numerical data and unprocessed blots are provided in Source Data Ext. Figure 2.
Extended Data Fig. 3 ZNF827 is critical for cellular migration without affecting cell viability and proliferation.
(a) Representative brightfield images of Scratch assay showing area filled after 18 hours of scratch by MDA-MB-231 cells transfected with control or ZNF827-specific siRNA. Data represent three biological replicates. (b-c) RT-PCR measurements of mRNA levels of key EMT relevant downregulated (b) and upregulated (c) genes in control and ZNF827 knockdown (4 days) MDA-MB-231 cells. RNA levels are shown relative to CTCF mRNA levels. (d) RT-PCR measurements of ZNF827 mRNA levels in MDA-MB-231 cells stably expressing either control shRNA or shRNA against ZNF827. mRNA levels are shown relative to CTCF mRNA levels. (e-f) Flow cytometry analysis of Annexin V (e) and BrdU (f) staining in control or ZNF827 depleted MDA-MB-231 cells (g-h) Kaplan-Meier analysis was performed for the Breast tumor datasets. Survival curves showing the overall survival in TCGA (g) and Breast invasive carcinoma (h). Error bars represent the mean ± SEM of three independent biological replicates for d. Statistical analyses were performed using a two-tailed unpaired Student’s t-test *p < 0.05 and ****p < 0.0001. n= two independent biological replicates for b-c. Source numerical data are provided in Source Data Ext. Figure 3.
Extended Data Fig. 4 Developing mouse brain as a model system for developmental EMT.
(a) Graphical representation of developing mouse neo-cortex and participating key cell types. Black arrows indicate cells delaminating from the neuroepithelial layer and migrating towards the cortical plate using apical radial glial cells as a scaffold. (b) The levels of key EMT markers are shown as normalized tag counts derived from the RNA-seq data obtained from the ventricular zone, the sub-ventricular zone, and the cortical plate of the E14.5 mouse cortex. (c) The levels of ZNF827 are shown as normalized tag counts derived from the RNA-seq data for distinct stages of in vitro neurogenesis, namely embryonic stem cells, neural progenitors and terminally differentiated neurons. (d) RT-PCR measurements of Zfp827 mRNA levels in FAC-sorted cells with markers specific for neuroepithelial cells (CD133) and cortical neurons (Ncam) from embryonic day 13.5 neocortex. mRNA levels for Znf827 are shown relative to Ctcf mRNA levels. Error bars represent the mean ± SEM of five independent biological replicates for b and three independent biological replicates for c-d. Statistical analyses were performed using a two-tailed unpaired Student’s t-test *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Source numerical data are provided in Source Data Ext. Figure 4.
Extended Data Fig. 5 ZNF827 is required for transcriptional changes critical for EMT.
(a) Schematic representation of Zinc finger domains of ZNF827. (b) Immunofluorescence image showing nuclear localization of ZNF827 in human primary breast epithelial (HMEC) and NMuMG cells. Scale bar: 20 and 100 μm respectively. (c) The mRNA levels of genes found to be downregulated and upregulated upon ZNF827 depletion during EMT were measured relative to CTCF via RT-PCRs (y-axis) in HMEC cells either untreated or undergoing TGF-β-induced EMT and treated with either non-targeting control (siControl) or siRNA against ZNF827 (siZNF827) (d) Representative bright-field images of HMLE cells untreated or after 4 days of TGF-β-induced EMT and siRNA-mediated depletion of ZNF827 with two independent siRNAs compared to the non-targeting control (siControl). (e) Volcano plot representation of RNA-seq results derived following ZNF827 knockdown in HMLE cells that underwent TGF-β-induced EMT for 4 days. The plot shows the number of genes with significantly altered expression along with GO analysis of downregulated and upregulated genes. (f) Venn diagram showing the overlap of deregulated genes from RNA-seq data following depletion of ZNF827 during EMT in HMEC and HMLE cells. (g) Representative bright-field images of HMLE cells after 4 days of TGF-β-induced EMT transfected with either a control vector or a construct overexpressing ZNF827. (h-i) Representative bright-field images of HMLE cells (h) and mRNA levels of key EMT markers (i) after 10 days of TGF-β-induced EMT and siRNA-mediated depletion of ZNF827 (siZNF827) compared to the non-targeting control (siControl). mRNA levels were measured relative to CTCF via RT-PCR, and the results were plotted on the y-axis. (j-k) Top non-redundant de novo (j) and known (k) DNA motifs enriched at the ZNF827 bound regions identified by HOMER. p values were generated by hypergeometric tests in HOMER. For b, d, g and h data represent three biological replicates. n = 2 independent biological replicates for c. Error bars represent the mean ± SEM of three independent biological replicates for i. Statistical analyses were performed using a two-tailed unpaired Student’s t-test *p < 0.05, and **p < 0.01. For e in volcano plot genes with significant differences (DESeq2, p-values cut-off of ≤0.05 attained by the Wald test are corrected for multiple testing using the Benjamini and Hochberg method, FDR < 5% from three independent biological replicates) and log2FC > 1 are highlighted in green (downregulated) or red (upregulated). The bars reflect the number of genes in each category, and other columns show significance of the enrichment (p-values) of a specific term determined using Fisher’s exact test and adjusted p-value using Benjamini correction. Source numerical data are provided in Source Data Ext. Figure 5.
Extended Data Fig. 6 ZNF827 depletion affects all types of splicing events during EMT.
(a-e) Scatterplot showing delta PSI for mutually exclusive exon (a), retained intron (b), alternative 3’ splice site (c), alternative 5’ splice site (d), and alternative last exon-AL (e), for genes during EMT (x-axis) and reverting to its epithelial state upon ZNF827 depletion (y-axis). The blue dots represent RNA-binding proteins. (f) Pie-chart showing overlap of genes annotated as splicing regulators and transcriptionally changing genes among the genes altered upon ZNF827 depletion during EMT. (g) Pie-chart showing the number of RNA interacting factors, undergoing alternative splicing in ZNF827-dependent manner during EMT. (h) A representative gene example displaying the sashimi plots showing transcriptional dynamics of a skipping exon during EMT and upon ZNF827 depletion. (i) Pie-chart showing overlap of genes annotated as splicing regulators and bound by ZNF827. (j) Pie-chart showing ZNF827 bound genes undergoing different types of splicing upon ZNF827 depletion during EMT. (k) RNA-seq data showing mRNA levels of FN1 in HMLE cells undergoing TGF-β-induced EMT; the y-axis represents normalized tag counts. (l-m) Splicing-PCR was performed using primer sets specific for different parts of the FN1 gene to measure skipping exon levels in HMLE cells either untreated or undergoing TGF-β-induced EMT and treated with either non-targeting control (siControl) or siRNA against ZNF827 (siZNF827) (l) as well as upon overexpression of ZNF827 (m). Three biological replicates are shown.
Extended Data Fig. 7 ZNF827-mediated splicing of QKI is critical for EMT progression.
(a) Quantification of QKI splicing isoform based on RNA-seq data during EMT and its reversal upon ZNF827 depletion. (b) Amino acid sequences of five major QKI protein isoforms show constant and variable amino acid residues. (c) Representative (n= two biological replicates) immunoblot showing expression dynamics of QKI in HMLE cells either untreated or undergoing TGF-β-induced EMT and treated with either non-targeting control (siControl) or siRNA against ZNF827 (siZNF827). GAPDH acts as a loading control. (d) Representative immuno-fluorescence images showing localization and expression levels of QKI and the mesenchymal marker Phalloidin (to visualize the actin cytoskeleton) in HMLE cells at 4 days post-transfection with two splicing isoforms of QKI. Data represent three biological replicates. (e) The levels of Zfp827 and Qk are shown as average normalized tag counts derived from the RNA-seq data (n = 3) from distinct stages of the in vitro neurogenesis, namely embryonic stem cells, neural progenitors, early neurons, and late neurons. The y-axis represents the average RPKM. (f) Representative DNA gel showing expression dynamics of Qk splicing isoforms during in vitro neurogenesis. Data represent three biological replicates. (g) Representative DNA gel showing PCR amplifications for splicing changes in various genes in HMLE cells either untreated or undergoing TGF-β-induced EMT and treated with either non-targeting control (siControl) or siRNA against ZNF827 (siZNF827) with or without QKI-5 overexpression. Data represent three biological replicates. (h) A Venn diagram showing the overlap of ZNF827-dependent and QKI-dependent splicing changes in HMLE cells. ZNF827 dependent splicing data was generated by us in HMLE cell during day 4 of TGF-β induction while QKi-dependent splicing data was generated following QKI overexpression in HMLE cells and collected from a previously published study. Source unprocessed blots are provided in Source Data Ext. Figure 7.
Extended Data Fig. 8 ZNF827 regulates the splicing of its target genes by modulating RNA Pol II processivity.
(a, b) Western blot analysis following immunoprecipitation assays to validate ZNF827 and HDAC1 interactions in HMLE (a) and NMuMG (b) undergoing TGF-β induced EMT. Data represent three biological replicates. (c) Splicing factors (in red) and RNA-binding proteins (in blue) are highlighted among selected significantly enriched proteins in the data from immunoprecipitation assay for ZNF827 in MDA-MB-231 cells followed by SILAC mass-spectrometry. (d) GO analysis of proteins enriched in the data from immunoprecipitation assay for ZNF827 in MDA-MB-231 cells followed by SILAC mass-spectrometry. The bars reflect the number of genes in each category, and the lines represent the significance of the enrichment (p-values) of a specific term determined using Fisher’s exact test displayed as an alternate x-axis of the corresponding GO terms. (e) Western blot analysis of immunoprecipitated samples to validate ZNF827 and U2AF2 interaction in HEK293 cells transfected with Flag-HA-ZNF827 and Myc-U2AF2. On the left IP for U2AF2 (anti-Myc) was performed and immunoblot was run to detect ZNF827 (anti-Flag). On the right, IP for ZNF827 (anti-Flag) was performed and immunoblot was performed to detect U2AF2 (anti-Myc). Data represent two biological replicates. (f) Immunofluorescence images showing nuclear co-localization of ZNF827 to U2AF1/U2AF2 in HEK293 cells. Data represent two biological replicates. (g) Box plot showing average nascent transcript signal for all genes bound by ZNF827 in HMLE cells undergoing TGF-β-induced EMT and treated with either non-targeting control (siControl) or siRNA against ZNF827 (siZNF827). In the box-and-whisker plots, the horizontal lines mark the median, the box limits indicate the 25th and 75th percentiles, and the whiskers extend to 1.5× the interquartile range from the 25th and 75th percentiles. The significance test was performed using two-tailed unpaired Wilcoxon test. (h-i) The average signal density of nascent transcript enrichment on gene body calculated by MACS for all genes bound by ZNF827 (h) and their average signal represented by box plot (i), of transcript in HMLE cells which are undergoing TGF-β-induced EMT and treated with either non-targeting control (siControl) or siRNA against ZNF827 (siZNF827). In the box-and-whisker plots, the horizontal lines mark the median, the box limits indicate the 25th and 75th percentiles, and the whiskers extend to 1.5× the interquartile range from the 25th and 75th percentiles. Two-tailed unpaired Wilcoxon test was used to calculate the P values. (j-k) Kinetics of RNA Pol II-dependent transcription elongation was measured in control and ZNF827 knockdown HMLE cells during EMT, and treated with DRB, and post-removal as described previously37. Quantitative real-time RT-PCRs were performed using primer sets specific for different parts of the indicated genes, RBM23 (j), and PCGF3 (k), to measure the levels of pre-mRNA expression. Primers were designed at the Exon-Intron junction or Intron-Exon Junction of pre-mRNAs towards 5’ ends, around middle, and around 3’ ends of each pre-mRNA. Camptothecin was used to block type I DNA topoisomerase and by doing so, it blocks transcription. Error bars represent the mean ± SEM of three independent biological replicates. Statistical analyses were performed using a two-tailed unpaired Student’s t-test *p < 0.05. (l) Representative bright-field images of three biological replicates showing phenotypic changes in HMLE cells which are either untreated or undergoing TGF-β-induced EMT and treated with either non-targeting control (siControl) with and without the HDAC inhibitor Sodium-butyrate or siRNA against ZNF827 (siZNF827). (m) Representative DNA gel showing PCR amplified AF in FIS1 in the same conditions described in (l). Data represent three biological replicates. (n) Representative DNA gel showing expression dynamics of QKI splicing isoforms (upper penal) and the box plot showing its quantification (lower penal) in HMLE cells which are either untreated or undergoing TGF-β-induced EMT and treated with either non-targeting control (siControl) or siRNA against ZNF827 (siZNF827) with and without HDAC and p300 inhibitors. Error bars represent the mean ± SEM of three independent biological replicates. Statistical analyses were performed using a two-tailed unpaired Student’s t-test *p < 0.05 Source numerical data and unprocessed blots are provided in Source Data Ext. Figure 8.
Supplementary information
Supplementary Tables 1–7
Table 1. Nucleotide sequence of primers used for RT–PCR, ChIP–PCR and splicing PCR. Nucleotide sequence of used siRNA. Table 2. a,b, Based on DESeq2, list of significantly (P-value cut-off of ≤0.05 attained by the Wald test corrected for multiple testing using the Benjamini and Hochberg method, FDR <5% from two biological replicates) 1.5-fold downregulated (a), and upregulated (b) genes in HMEC cells upon siRNA-mediated depletion of ZNF827. c,d, Based on DESeq2, list of significantly (P-value cut-off of ≤0.05 attained by the Wald test corrected for multiple testing using the Benjamini and Hochberg method, FDR <5% from two biological replicates) de-regulated genes in HMLE cells during EMT (c) and upon siRNA-mediated depletion of ZNF827 (d). Table 3. a,b, List of ZNF827 bound ChIP (a) and CUT&Tag (b) peaks. n = two independent biological replicates. Table 4. a–c, List of significantly (DESeq2, P-value cut-off of ≤0.05 attained by the Wald test corrected for multiple testing using the Benjamini and Hochberg method, FDR <5% from three biological replicates) de-regulated spliced isoform in HMLE cells during EMT (a), upon siRNA-mediated depletion of ZNF827 (b) and reverting towards epithelial cell (c). Table 5. QKI-dependent splicing changes and its comparison with ZNF827 dependent splicing changes. Table 6: ZNF827-dependent splicing changes harbouring QKI motifs. Table 7. ZNF827 interactors based on SILAC massspec.
Supplementary Data
Vector map for all used plasmids.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed western blots and/or gels.
Source Data Fig. 8
Statistical source data.
Source Data Fig. 8
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots and/or gels.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sahu, S.K., Agirre, E., Inayatullah, M. et al. A complex epigenome-splicing crosstalk governs epithelial-to-mesenchymal transition in metastasis and brain development. Nat Cell Biol 24, 1265–1277 (2022). https://doi.org/10.1038/s41556-022-00971-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-00971-3
This article is cited by
-
Transcription factors in fibroblast plasticity and CAF heterogeneity
Journal of Experimental & Clinical Cancer Research (2023)
-
The epithelial–mesenchymal plasticity landscape: principles of design and mechanisms of regulation
Nature Reviews Genetics (2023)
-
Cancer metastasis under the magnifying glass of epigenetics and epitranscriptomics
Cancer and Metastasis Reviews (2023)