Abstract
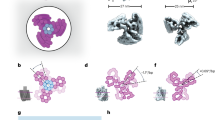
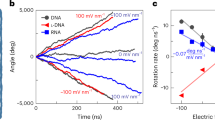
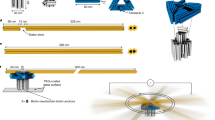
Flow-driven rotary motors such as windmills and water wheels drive functional processes in human society. Although examples of such rotary motors also feature prominently in cell biology, their synthetic construction at the nanoscale has remained challenging. Here we demonstrate flow-driven rotary motion of a self-organized DNA nanostructure that is docked onto a nanopore in a thin solid-state membrane. An elastic DNA bundle self-assembles into a chiral conformation upon phoretic docking onto the solid-state nanopore, and subsequently displays a sustained unidirectional rotary motion of up to 20 rev s−1. The rotors harness energy from a nanoscale water and ion flow that is generated by a static chemical or electrochemical potential gradient in the nanopore, which are established through a salt gradient or applied voltage, respectively. These artificial nanoengines self-organize and operate autonomously in physiological conditions, suggesting ways to constructing energy-transducing motors at nanoscale interfaces.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All experimental data are available at https://doi.org/10.5281/zenodo.6513594.
Code availability
MATLAB codes for data processing are available at https://doi.org/10.5281/zenodo.6513594. Julia codes used for numerical simulation are available at https://gitlab.gwdg.de/LMP-pub/nanoturbines.
References
Yoshida, M., Muneyuki, E. & Hisabori, T. ATP synthase—a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677 (2001).
Srivastava, A. P. et al. High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane. Science 360, eaas9699 (2018).
Minamino, T., Imada, K. & Namba, K. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 18, 693–701 (2008).
Perkins, G. et al. Electron tomography of neuronal mitochondria: three-dimensional structure and organization of cristae and membrane contacts. J. Struct. Biol. 119, 260–272 (1997).
Ramezani, H. & Dietz, H. Building machines with DNA molecules. Nat. Rev. Genet. 21, 5–26 (2020).
Ozin, G. A., Manners, I., Fournier‐Bidoz, S. & Arsenault, A. Dream nanomachines. Adv. Mater. 17, 3011–3018 (2005).
Browne, W. & Feringa, B. Making molecular machines work. Nat. Nanotechnol. 1, 25–35 (2006).
Astumian, R. D. Thermodynamics and kinetics of a Brownian motor. Science 276, 917–922 (1997).
Feynman, R. There’s plenty of room at the bottom: an invitation to open up a new field of physics. Eng. Sci. 23, 22–36 (1960).
Brown, A. I. & Sivak, D. A. Theory of nonequilibrium free energy transduction by molecular machines. Chem. Rev. 120, 434–459 (2019).
Stoddart, J. F. The chemistry of the mechanical bond. Chem. Soc. Rev. 38, 1802–1820 (2009).
García-López, V., Liu, D. & Tour, J. M. Light-activated organic molecular motors and their applications. Chem. Rev. 120, 79–124 (2019).
Kopperger, E. et al. A self-assembled nanoscale robotic arm controlled by electric fields. Science 359, 296–301 (2018).
Thubagere, A. J. et al. A cargo-sorting DNA robot. Science 357, eaan6558 (2017).
Soong, R. K. et al. Powering an inorganic nanodevice with a biomolecular motor. Science 290, 1555–1558 (2000).
Yehl, K. et al. High-speed DNA-based rolling motors powered by RNase H. Nat. Nanotechnol. 11, 184–190 (2016).
Golestanian, R. Synthetic mechanochemical molecular swimmer. Phys. Rev. Lett. 105, 018103 (2010).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Van Dorp, S., Keyser, U. F., Dekker, N. H., Dekker, C. & Lemay, S. G. Origin of the electrophoretic force on DNA in solid-state nanopores. Nat. Phys. 5, 347–351 (2009).
Anderson, J. L. Colloid transport by interfacial forces. Annu. Rev. Fluid Mech. 21, 61–99 (1989).
Golestanian, R. Phoretic active matter. Preprint at https://arxiv.org/abs/1909.03747 (2019).
Castro, C. E., Su, H.-J., Marras, A. E., Zhou, L. & Johnson, J. Mechanical design of DNA nanostructures. Nanoscale 7, 5913–5921 (2015).
Bergou, M., Wardetzky, M., Robinson, S., Audoly, B. & Grinspun, E. Discrete elastic rods. ACM Trans. Graph 27, 1–12 (2008).
Lee, C. et al. Osmotic flow through fully permeable nanochannels. Phys. Rev. Lett. 112, 244501 (2014).
Bonthuis, D. J. & Golestanian, R. Mechanosensitive channel activation by diffusio-osmotic force. Phys. Rev. Lett. 113, 148101 (2014).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Wagenbauer, K. F. et al. How we make DNA origami. ChemBioChem 18, 1873–1885 (2017).
Verschueren, D. V., Yang, W. & Dekker, C. Lithography-based fabrication of nanopore arrays in freestanding SiN and graphene membranes. Nanotechnology 29, 145302 (2018).
Janssen, X. J. et al. Rapid manufacturing of low-noise membranes for nanopore sensors by trans-chip illumination lithography. Nanotechnology 23, 475302 (2012).
Churchman, L. S., Ökten, Z., Rock, R. S., Dawson, J. F. & Spudich, J. A. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc. Natl Acad. Sci. USA 102, 1419–1423 (2005).
Kwok, H., Briggs, K. & Tabard-Cossa, V. Nanopore fabrication by controlled dielectric breakdown. PLoS ONE 9, e92880 (2014).
Ovesný, M., Křížek, P., Borkovec, J., Švindrych, Z. & Hagen, G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Lyubchenko, Y. L. & Shlyakhtenko, L. S. AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods 47, 206–213 (2009).
Horcas, I. et al. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 78, 013705 (2007).
Acknowledgements
We thank A. Aksimentiev, C. Maffeo, M. Tišma, A. Fragasso and A. Barth for discussions, B. Pradhan for help with the single-molecule fluorescence set-up, N. Klughammer for help with the fabrication of fiducial marker grids for dual-channel fluorescence imaging, and P. Ketterer for initial DNA origami structure designs. We acknowledge funding support by Dutch Research Council NWO grant no. NWO-I680 and the European Research Council Advanced Grant 883684 (C.D.). This work was supported by a European Research Council Consolidator Grant to H.D. (GA no. 724261), the Deutsche Forschungsgemeinschaft through grants provided within the Gottfried-Wilhelm-Leibniz Program (H.D.), and the SFB863 Project ID 111166240 TPA9 (H.D.). The work has received support from the Max Planck School Matter to Life (R.G. and H.D.) and the MaxSynBio Consortium (R.G.), which are jointly funded by the Federal Ministry of Education and Research (BMBF) of Germany and the Max Planck Society.
Author information
Authors and Affiliations
Contributions
X.S., D.V., H.D. and C.D. conceived the concept of DNA rotors in nanopores. A.-K.P. and H.D. designed and prepared the DNA origami structures. X.S. designed the nanopore experiment and fabricated nanopore devices. X.S. and W.Z. conducted nanopore experiments. A.M.-G. performed AFM measurements. X.S. and D.V. wrote the data analysis program and analysed data. J.I. and R.G. designed and conducted theoretical modelling and simulations. All authors discussed the experimental findings and co-wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Physics thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–4, Figs. 1–14, Tables 1–5 and captions for Videos 1–6.
Supplementary Table
DNA sequences of staple strands
Supplementary Video 1
Rotary motion of DNA rotors on nanopores. Top row: raw video of the Cy5 channel of each rotor. Middle row: corresponding single-particle localization results of both ends of the DNA rotors. The position of the current frame is marked as orange and blue dots, and the trajectory of the 10 frames before the current frame is shown as solid lines. The two dots are connected with a red bar. Bottom row: corresponding cumulative angular displacement (𝑡). All plots in the video are synced. The video playback frame rate is 40 fps, which is around 10 times slower than the original data (450–500 fps).
Supplementary Video 2
Simulated DNA rotors on nanopores. Top view of a collection of different simulated DNA rotors. The 6hb rods are shown in orange and the rim of the pore in red. All parameters were the same for each panel in this video, except for the initial placement of the 6hb rod (shown in blue) on the nanopore. Simulations that terminated early due to translocation of the rod through the pore are marked by a grey background.
Supplementary Video3
Example (1) of bending configurations in 3D simulations of DNA rods on nanopores. A portion of the membrane is shown in grey, the rim of the pore is highlighted in red, and a 3D rendering of the motion of the DNA rod is displayed.
Supplementary Video 4
Example (2) of bending configurations in 3D simulations of DNA rods on nanopores. A portion of the membrane is shown in grey, the rim of the pore is highlighted in red, and a 3D rendering of the motion of the DNA rod is displayed.
Supplementary Video 5
Example (3) of bending configurations in 3D simulations of DNA rods on nanopores. A portion of the membrane is shown in grey, the rim of the pore is highlighted in red, and a 3D rendering of the motion of the DNA rod is displayed.
Supplementary Video 6
Example (4) of bending configurations in 3D simulations of DNA rods on nanopores. A portion of the membrane is shown in grey, the rim of the pore is highlighted in red, and a 3D rendering of the motion of the DNA rod is displayed.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, X., Pumm, AK., Isensee, J. et al. Sustained unidirectional rotation of a self-organized DNA rotor on a nanopore. Nat. Phys. 18, 1105–1111 (2022). https://doi.org/10.1038/s41567-022-01683-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-022-01683-z
This article is cited by
-
A rhythmically pulsing leaf-spring DNA-origami nanoengine that drives a passive follower
Nature Nanotechnology (2024)
-
Nanoturbine driven by flow across a nanopore
Nature Nanotechnology (2024)
-
A DNA turbine powered by a transmembrane potential across a nanopore
Nature Nanotechnology (2024)
-
Ratcheting synthesis
Nature Reviews Chemistry (2023)
-
Nanopores enable electrohydrodynamical DNA motor
Nature Nanotechnology (2023)