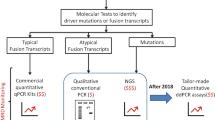
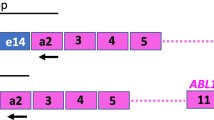
Abstract
One of the indications for BCR::ABL1 mutation testing in chronic myeloid leukemia (CML) is when tyrosine kinase inhibitor therapy (TKI) needs to be changed for unsatisfactory response. In this study, we evaluated a droplet digital PCR (ddPCR)-based multiplex strategy for the detection and quantitation of transcripts harbouring mutations conferring resistance to second-generation TKIs (2GTKIs). Parallel quantitation of e13a2, e14a2 and e1a2 BCR::ABL1 fusion transcripts enables to express results as percentage of mutation positive- over total BCR::ABL1 transcripts. We determined the limit of blank in 60 mutation-negative samples. Accuracy was demonstrated by further analysis of 48 samples already studied by next generation sequencing (NGS). Mutations could be called down to 0.5% and across 3-logs of BCR::ABL1 levels. Retrospective review of BCR::ABL1 NGS results in 513 consecutive CML patients with non-optimal response to first- or second-line TKI therapy suggested that a ddPCR-based approach targeted against 2GTKI-resistant mutations would score samples as mutation-negative in 22% of patients with warning response to imatinib but only in 6% of patients with warning response to 2GTKIs. We conclude ddPCR represents an attractive method for easy, accurate and rapid screening for 2GTKI-resistant mutations impacting on TKI selection, although ddPCR cannot identify compound mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Materials described in this manuscript, including all relevant raw data, will be freely available upon request to any researcher wishing to use them for non-commercial purposes.
References
Bavaro L, Martelli M, Cavo M, Soverini S. Mechanisms of Disease Progression and Resistance to Tyrosine Kinase Inhibitor Therapy in Chronic Myeloid Leukemia: An Update. Int J Mol Sci. 2019;20. https://doi.org/10.3390/ijms20246141
Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020;34:966–84. https://doi.org/10.1038/s41375-020-0776-2
Cortes JE, Kantarjian H, Shah NP, Bixby D, Mauro MJ, Flinn I, et al. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med. 2012;367:2075–88. https://doi.org/10.1056/NEJMoa1205127
Hughes TP, Mauro MJ, Cortes JE, Minami H, Rea D, DeAngelo DJ, et al. Asciminib in Chronic Myeloid Leukemia after ABL Kinase Inhibitor Failure. N Engl J Med. 2019;381:2315–26. https://doi.org/10.1056/NEJMoa1902328
Soverini S, Hochhaus A, Nicolini FE, Gruber F, Lange T, Saglio G, et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: recommendations from an expert panel on behalf of European LeukemiaNet. Blood 2011;118:1208–15. https://doi.org/10.1182/blood-2010-12-326405
Soverini S, De Benedittis C, Machova Polakova K, Brouckova A, Horner D, Iacono M, et al. Unraveling the complexity of tyrosine kinase inhibitor-resistant populations by ultra-deep sequencing of the BCR-ABL kinase domain. Blood 2013;122:1634–48. https://doi.org/10.1182/blood-2013-03-487728
Machova Polakova K, Kulvait V, Benesova A, Linhartova J, Klamova H, Jaruskova M, et al. Next-generation deep sequencing improves detection of BCR-ABL1 kinase domain mutations emerging under tyrosine kinase inhibitor treatment of chronic myeloid leukemia patients in chronic phase. J Cancer Res Clin Oncol. 2015;141:887–99. https://doi.org/10.1007/s00432-014-1845-6
Baer C, Kern W, Koch S, Nadarajah N, Schindela S, Meggendorfer M, et al. Ultra-deep sequencing leads to earlier and more sensitive detection of the tyrosine kinase inhibitor resistance mutation T315I in chronic myeloid leukemia. Haematologica 2016;101:830–8. https://doi.org/10.3324/haematol.2016.145888
Soverini S, De Benedittis C, Polakova KM, Linhartova J, Castagnetti F, Gugliotta G, et al. Next-generation sequencing for sensitive detection of BCR-ABL1 mutations relevant to tyrosine kinase inhibitor choice in imatinib-resistant patients. Oncotarget 2016;7:21982–90. https://doi.org/10.18632/oncotarget.8010
Soverini S, De Benedittis C, Castagnetti F, Gugliotta G, Mancini M, Bavaro L, et al. In chronic myeloid leukemia patients on second-line tyrosine kinase inhibitor therapy, deep sequencing of BCR-ABL1 at the time of warning may allow sensitive detection of emerging drug-resistant mutants. BMC Cancer. 2016;16:572 https://doi.org/10.1186/s12885-016-2635-0
Kizilors A, Crisa E, Lea N, Passera R, Mian S, Anwar J, et al. Effect of low-level BCR-ABL1 kinase domain mutations identified by next-generation sequencing in patients with chronic myeloid leukaemia: A population-based study. Lancet Haematol. 2019;6:e276–e84. https://doi.org/10.1016/S2352-3026(19)30027-4
Soverini S, Bavaro L, De Benedittis C, Martelli M, Iurlo A, Orofino N, et al. Prospective assessment of NGS-detectable mutations in CML patients with nonoptimal response: the NEXT-in-CML study. Blood 2020;135:534–41. https://doi.org/10.1182/blood.2019002969
Zabriskie MS, Eide CA, Tantravahi SK, Vellore NA, Estrada J, Nicolini FE, et al. BCR-ABL1 compound mutations combining key kinase domain positions confer clinical resistance to ponatinib in Ph chromosome-positive leukemia. Cancer Cell. 2014;26:428–42. https://doi.org/10.1016/j.ccr.2014.07.006
Byrgazov K, Lucini CB, Valent P, Hantschel O, Lion T. BCR-ABL1 compound mutants display differential and dose-dependent responses to ponatinib. Haematologica 2018;103:e10–e2. https://doi.org/10.3324/haematol.2017.176347
Soverini S, Martelli M, Bavaro L, De Benedittis C, Sica S, Sora F, et al. BCR-ABL1 compound mutants: prevalence, spectrum and correlation with tyrosine kinase inhibitor resistance in a consecutive series of Philadelphia chromosome-positive leukemia patients analyzed by NGS. Leukemia 2021;35:2102–7. https://doi.org/10.1038/s41375-020-01098-w
Flach J, Shumilov E, Joncourt R, Porret N, Novak U, Pabst T, et al. Current concepts and future directions for hemato-oncologic diagnostics. Crit Rev Oncol Hematol. 2020;151:102977 https://doi.org/10.1016/j.critrevonc.2020.102977
Olmedillas-Lopez S, Olivera-Salazar R, Garcia-Arranz M, Garcia-Olmo D. Current and Emerging Applications of Droplet Digital PCR in Oncology: An Updated Review. Mol Diagn Ther. 2022;26:61–87. https://doi.org/10.1007/s40291-021-00562-2
Huggett JF. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin Chem. 2020;66:1012–29. https://doi.org/10.1093/clinchem/hvaa125
Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, Apperley JF, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 2013;122:872–84. https://doi.org/10.1182/blood-2013-05-501569
Soverini S, Abruzzese E, Bocchia M, Bonifacio M, Galimberti S, Gozzini A, et al. Next-generation sequencing for BCR-ABL1 kinase domain mutation testing in patients with chronic myeloid leukemia: a position paper. J Hematol Oncol. 2019;12:131 https://doi.org/10.1186/s13045-019-0815-5
Regan JF, Kamitaki N, Legler T, Cooper S, Klitgord N, Karlin-Neumann G, et al. A rapid molecular approach for chromosomal phasing. PLoS One. 2015;10:e0118270 https://doi.org/10.1371/journal.pone.0118270
Vannuffel P, Bavaro L, Nollet F, Aynaci A, Martelli M, Devos H, et al. Droplet Digital PCR Phasing (DROP-PHASE): A Novel Method for Straightforward Detection of BCR-ABL1 Compound Mutations in Tyrosine Kinase Inhibitors Resistant Chronic Myeloid Leukemia (CML) and Acute Lymphoblastic Leukemia (ALL). Blood 2019;134:4660.
Soverini S, De Benedittis C, Papayannidis C, Polakova KM, Venturi C, Russo D, et al. Clinical impact of low-burden BCR-ABL1 mutations detectable by amplicon deep sequencing in Philadelphia-positive acute lymphoblastic leukemia patients. Leukemia 2016;30:1615–9. https://doi.org/10.1038/leu.2016.17
Acknowledgements
The study was supported by Novartis through the European Treatment and Outcome Study (EUTOS) for CML. ddPCR assays were provided by Bio-Rad Laboratories.
Author information
Authors and Affiliations
Contributions
SS: designed the study, analyzed and interpreted results, contributed to writing the report; SDS, MMartelli, CM, SB, MMancini, CV: performed the study, contributed to data analysis and interpretation, prepared the figures and tables; FC, GG, CP: performed patient and sample selection, contributed to data analysis and interpretation; KMP, TE: provided important study materials, contributed to data analysis and interpretation, contributed to writing the report; DM, AC: provided reagents, contributed to experimental protocol set up and optimization; MC, designed the study, coordinated the clinical and research groups, contributed to writing the report. All authors read and approved the manuscript for submission.
Corresponding author
Ethics declarations
Competing interests
ddPCR assays for this study were provided by Bio-Rad Laboratories. Bio-Rad Laboratories had no role in the design of the study, collection and analysis of data and decision to publish. DM and AC are Bio-Rad’s employees. The remaining authors declare no conflicts of interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Soverini, S., De Santis, S., Martelli, M. et al. Droplet digital PCR for the detection of second-generation tyrosine kinase inhibitor-resistant BCR::ABL1 kinase domain mutations in chronic myeloid leukemia. Leukemia 36, 2250–2260 (2022). https://doi.org/10.1038/s41375-022-01660-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01660-8