Abstract
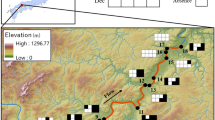
Invasive aquatic plants can have various ecological impacts on river ecosystems. It is important to identify the spatial positions of the invasive aquatic plant sources in a mainstem-tributary network to understand the distributions and prioritize the removal areas of invasive aquatic plants. This study used environmental DNA (eDNA) analyses to identify the potential sources (invasive macrophyte patches that persist throughout seasons in a river segment and can provide the plant fragments to the downstream) of Egeria densa, an invasive aquatic macrophyte, in a riverine network. We monitored the seasonal changes (March, July, August, and October) of eDNA concentrations of E. densa in tributaries with their associated mainstem sites (total 16 × 2 = 32 sites) in the Gonokawa River, Japan in 2021 and identified the potential sources of the target species based on the spatial abundance patterns and persistence in the riverine network. Our results indicated that the eDNA concentrations of E. densa have decreased in the mainstem from March to October, whereas the eDNA concentrations in the tributary sites had opposite temporal patterns possibly because the artificial tributary sites could be less affected by the flood event. In addition, certain mainstem segments and tributaries had higher eDNA concentrations and persistence of E. densa than other locations and could be potential sources of the invasive macrophyte in the system. Our study helps to identify and remove the potential sources of invasive macrophytes and prevent the further spread in riverine systems.








Similar content being viewed by others
References
Anglès d’Auriac MB, Strand DA, Mjelde M, Demars BOL, Thaulow J (2019) Detection of an invasive aquatic plant in natural water bodies using environmental DNA. PLoS One. https://doi.org/10.1371/journal.pone.0219700
Conrad JL, Bibian AJ, Weinersmith KL, Carion DD, Young MJ, Crain P, Hestir EL, Santos MJ, Sih A (2016) Novel species interactions in a highly modified estuary: association of largemouth bass with brazzilian watershed Egeria densa. Trans Am Fish Soc 145:249–263. https://doi.org/10.1080/00028487.2015.1114521
Crooks JA (2002) Characterizing ecosystem-level consequences of biological invasions: the role of ecosystem engineers. Oikos 97:153–166. https://doi.org/10.1034/j.1600-0706.2002.970201.x
Doi H, Akamatsu Y, Goto M, Inui R, Komuro T, Nagano M, Minamoto T (2021) Broad-scale detection of environmental DNA for an invasive macrophyte and the relationship between DNA concentration and coverage in rivers. Biol Invasions 23:507–520. https://doi.org/10.1007/s10530-020-02380-9
Feijoó CS, Momo FR, Benetto CA, Tur NM (1996) Factors influencing biomass and nutrient content of the submersed macrophyte Egeria densa Planch. in a pampasic stream. Hydrobiologia 341:21–26
Fujiwara A, Matsuhashi S, Doi H, Yamamoto S, Minamoto T (2016) Use of environmental DNA to survey the distribution of an invasive submerged plant in ponds. Freshw Sci 35:748–754. https://doi.org/10.1086/685882
Gantz CA, Renshaw MA, Erickson D, Lodge DM, Egan SP (2018) Environmental DNA detection of aquatic invasive plants in lab mesocosm and natural field conditions. Biol Invasions 20:2535–2552. https://doi.org/10.1007/s10530-018-1718-z
Haramoto T, Ikusima I (1988) Life cycle of Egeria densa Planch., an aquatic plant naturalized in Japan. Aquat Bot 30:389–403. https://doi.org/10.1016/0304-3770(88)90070-8
Heidbüchel P, Hussner A (2020) Falling into pieces: In situ fragmentation rates of submerged aquatic plants and the influence of discharge in lowland streams. Aquat Bot 160:103–164. https://doi.org/10.1016/j.aquabot.2019.103164
Kadono Y (2004) Alien aquatic plants naturalized in Japan: history and present status. Glob Environ Res 8:163–169
Kodama T, Akamatsu Y, Miyazono S, Yamaguchi K (2019) Examination of washout characteristic of Egeria densa during large flood. J Jpn Soc Civ Eng Ser B1 (hydraulic Engineering) 75:391–396. https://doi.org/10.2208/jscejhe.75.2_I_391 (in Japanese)
Kodama T, Miyazono S, Akamatsu Y, Yamaguchi K (2020) Examination of environmental factors affecting the large spatial distribution of Egeria densa. J Jpn Soc Civ Eng Ser Ser B1 (hydraulic Engineering) 76:1261–1266 (in Japanese)
Kodama T, Miyazono S, Akamatsu Y (2021) Elucidation of environmental factors affecting the settlement of Egeria densa in the downstream segment of the Haji Dam. Jpn Soc Civ Eng Ser Ser B1 (hydraulic Engineering) 77:973–978 (in Japanese)
Kodama T, Miyazono S, Akamatsu Y, Tsuji S, Nakao R (2022) Abundance estimation of riverine macrophyte Egeria densa using environmental DNA: effects of sampling season and location. Limnol 23:299–308
Kralik P, Ricchi M (2017) A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol 8:108. https://doi.org/10.3389/fmicb.2017.00108
Kuehne LM, Ostberg CO, Chase DM, Duda JJ, Olden JD (2020) Use of environmental DNA to detect the invasive aquatic plants Myriophyllum spicatum and Egeria densa in lakes. Freshw Sci 39:521–533. https://doi.org/10.1086/710106
Lund EM, Drake DC (2021) Species-specific wet-dry mass calibrations for common submersed macrophytes in the Upper Mississippi River. Aquat Bot 169:103344
Matsuhashi S, Minamoto T, Doi H (2019) Seasonal change in environmental DNA concentration of a submerged aquatic plant species. Freshw Sci 38:654–660. https://doi.org/10.1086/704996
Miyazono S, Taylor CM (2013) Effects of habitat size and isolation on species immigration-extinction dynamics and community nestedness in a desert river system. Freshw Biol 58:1303–1312
Miyazono S, Kodama T, Akamatsu Y, Nakao R, Saito M (2021) Application of environmental DNA methods for the detection and abundance estimation of invasive aquatic plant Egeria densa in lotic habitats. Limnol 22:81–87. https://doi.org/10.1007/s10201-020-00636-w
Miyazono S, Kodama T, Akamatsu Y, Nakao R, Saito M, Tsuji S (2022) Evaluation of the tributaries as habitats for Ayu in the Gonokawa River using environmental DNA analysis. Ecol Civ Eng 24:259–266 (in Japanese)
R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Tsuji S, Takahara T, Doi H, Shibata N, Yamanaka H (2019) The detection of aquatic macroorganisms using environmental DNA analysis —a review of methods for collection, extraction, and detection. Environmental DNA 1:99–108. https://doi.org/10.1002/edn3.21
Tsuji S, Nakao R, Saito M, Minamoto T, Akamatsu Y (2022) Pre-centrifugation before DNA extraction mitigates extraction efficiency reduction of environmental DNA caused by the preservative solution (benzalkonium chloride) remaining in the filters. Limnol 23:9–16. https://doi.org/10.1007/s10201-021-00676-w
Yamanaka H, Minamoto T, Matsuura J, Sakurai S, Tsuji S, Motozawa H, Hongo M, Sogo Y, Kakimi N, Teramura I, Sugita M, Baba M, Kondo A (2017) A simple method for preserving environmental DNA in water samples at ambient temperature by addition of cationic surfactant. Limnol 18:233–241. https://doi.org/10.1007/s10201-016-0508-5
Yarrow M, Marin VH, Finlayson M, Tironi A, Delgado LE, Fischer F (2009) The ecology of Egeria densa Planchon (Liliopsida: Alismatales): a wetland ecosystem engineer? Rev Chil Hist Nat 82:299–313. https://doi.org/10.4067/S0716-078X2009000200010
Acknowledgements
This research was supported by Chugoku Regional Development Bureau of Ministry of Land, Infrastructure, Transport and Tourism. We thank two anonymous reviewers for the helpful comments on our manuscript. The experiment reported here complied with current laws of Japan.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Miyazono, S., Kodama, T., Akamatsu, Y. et al. Application of environmental DNA analysis for detecting potential sources of invasive aquatic plant Egeria densa in a riverine network. Landscape Ecol Eng 19, 45–54 (2023). https://doi.org/10.1007/s11355-022-00517-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11355-022-00517-7