Abstract
Purpose
Deep learning is an emerging reconstruction method for positron emission tomography (PET), which can tackle complex PET corrections in an integrated procedure. This paper optimizes the direct PET reconstruction from sinogram on a long axial field of view (LAFOV) PET.
Methods
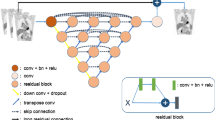
This paper proposes a novel deep learning architecture to reduce the biases during direct reconstruction from sinograms to images. This architecture is based on an encoder-decoder network, where the perceptual loss is used with pre-trained convolutional layers. It is trained and tested on data of 80 patients acquired from recent Siemens Biograph Vision Quadra long axial FOV (LAFOV) PET/CT. The patients are randomly split into a training dataset of 60 patients, a validation dataset of 10 patients, and a test dataset of 10 patients. The 3D sinograms are converted into 2D sinogram slices and used as input to the network. In addition, the vendor reconstructed images are considered as ground truths. Finally, the proposed method is compared with DeepPET, a benchmark deep learning method for PET reconstruction.
Results
Compared with DeepPET, the proposed network significantly reduces the root-mean-squared error (NRMSE) from 0.63 to 0.6 (p < 0.01) and increases the structural similarity index (SSIM) and peak signal-to-noise ratio (PSNR) from 0.93 to 0.95 (p < 0.01) and from 82.02 to 82.36 (p < 0.01), respectively. The reconstruction time is approximately 10 s per patient, which is shortened by 23 times compared with the conventional method. The errors of mean standardized uptake values (SUVmean) for lesions between ground truth and the predicted result are reduced from 33.5 to 18.7% (p = 0.03). In addition, the error of max SUV is reduced from 32.7 to 21.8% (p = 0.02).
Conclusion
The results demonstrate the feasibility of using deep learning to reconstruct images with acceptable image quality and short reconstruction time. It is shown that the proposed method can improve the quality of deep learning-based reconstructed images without additional CT images for attenuation and scattering corrections. This study demonstrated the feasibility of deep learning to rapidly reconstruct images without additional CT images for complex corrections from actual clinical measurements on LAFOV PET. Despite improving the current development, AI-based reconstruction does not work appropriately for untrained scenarios due to limited extrapolation capability and cannot completely replace conventional reconstruction currently.







Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Vandenberghe S, Moskal P, Karp JS. State of the art in total body PET. EJNMMI Phys. 2020;7:35. https://doi.org/10.1186/s40658-020-00290-2.
Cherry SR, Jones T, Karp JS, Qi J, Moses WW, Badawi RD. Total-body PET: maximizing sensitivity to create new opportunities for clinical research and patient care. J Nucl Med : Off Publ Soc Nucl Med. 2018;59:3–12. https://doi.org/10.2967/jnumed.116.184028.
Cherry SR, Badawi RD, Karp JS, Moses WW, Price P, Jones T. Total-body imaging: transforming the role of positron emission tomography. Sci Transl Med. 2017;9:eaaf6169. https://doi.org/10.1126/scitranslmed.aaf6169.
Defrise M, Kinahan PE, Michel CJ. Image reconstruction algorithms in PET. In: Bailey DL, Townsend DW, Valk PE, Maisey MN, editors. Positron emission tomography: basic sciences. London: Springer, London; 2005. p. 63–91.
Shepp LA, Vardi Y. Maximum likelihood reconstruction for emission tomography. IEEE Trans Med Imaging. 1982;1:113–22. https://doi.org/10.1109/TMI.1982.4307558.
Hudson HM, Larkin RS. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans Med Imaging. 1994;13:601–9. https://doi.org/10.1109/42.363108.
Uribe CF, Mathotaarachchi S, Gaudet V, Smith KC, Rosa-Neto P, Benard F, et al. Machine learning in nuclear medicine: part 1—introduction. J Nucl Med. 2019;60:451–8. https://doi.org/10.2967/jnumed.118.223495.
Reader AJ, Corda G, Mehranian A, da Costa-Luis C, Ellis S, Schnabel JA. Deep learning for PET image reconstruction. IEEE Trans Radiat Plasma Med Sci. 2020;1-. https://doi.org/10.1109/trpms.2020.3014786.
Gong K, Guan J, Liu CC, Qi J. PET image denoising using a deep neural network through fine tuning. IEEE Trans Radiat Plasma Med Sci. 2019;3:153–61. https://doi.org/10.1109/TRPMS.2018.2877644.
Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, et al. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging. 2019;46:2780–9.
Katsari K, Penna D, Arena V, Polverari G, Ianniello A, Italiano D, et al. Artificial intelligence for reduced dose 18F-FDG PET examinations: a real-world deployment through a standardized framework and business case assessment. EJNMMI Physics. 2021;8:25. https://doi.org/10.1186/s40658-021-00374-7.
Gong K, Guan J, Kim K, Zhang X, Fakhri G, Qi J, et al. Iterative PET image reconstruction using convolutional neural network representation. IEEE Trans Med Imaging. 2017;38. https://doi.org/10.1109/TMI.2018.2869871.
Gong K, Wu D, Kim K, Yang J, Sun T, El Fakhri G, et al. MAPEM-Net: an unrolled neural network for fully 3D PET image reconstruction. 15th International Meeting on Fully Three-Dimensional Image Reconstruction in Radiology and Nuclear Medicine: Int Soc Opt Photon. 2019; p. 110720O.
Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature. 2018;555:487–92. https://doi.org/10.1038/nature25988.
Haggstrom I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: a deep encoder-decoder network for directly solving the PET image reconstruction inverse problem. Med Image Anal. 2019;54:253–62. https://doi.org/10.1016/j.media.2019.03.013.
Kandarpa VSS, Bousse A, Benoit D, Visvikis D. DUG-RECON: a framework for direct image reconstruction using convolutional generative networks. IEEE Trans Radiat Plasma Med Sci. 2021;5:44–53. https://doi.org/10.1109/trpms.2020.3033172.
Whiteley W, Luk WK, Gregor J. DirectPET: full-size neural network PET reconstruction from sinogram data. J Med Imaging (Bellingham). 2020;7:032503. https://doi.org/10.1117/1.JMI.7.3.032503.
Whiteley W, Panin V, Zhou C, Cabello J, Bharkhada D, Gregor J. FastPET: near real-time PET reconstruction from histo-images using a neural network. arXiv preprint arXiv:200204665. 2020.
Schmall JP, Karp JS, Werner M, Surti S. Parallax error in long-axial field-of-view PET scanners—a simulation study. Phys Med Biol. 2016;61:5443–55. https://doi.org/10.1088/0031-9155/61/14/5443.
Zhang X, Badawi RD, Cherry SR, Qi J. Theoretical study of the benefit of long axial field-of-view PET on region of interest quantification. Phys Med Biol. 2018;63:135010. https://doi.org/10.1088/1361-6560/aac815.
Efthimiou N. New challenges for PET image reconstruction for total-body imaging. PET Clin. 2020;15:453–61. https://doi.org/10.1016/j.cpet.2020.06.002.
Zhang X, Zhou J, Cherry SR, Badawi RD, Qi J. Quantitative image reconstruction for total-body PET imaging using the 2-meter long EXPLORER scanner. Phys Med Biol. 2017;62:2465–85. https://doi.org/10.1088/1361-6560/aa5e46.
Alberts I, Hünermund JN, Prenosil G, Mingels C, Bohn KP, Viscione M, et al. Clinical performance of long axial field of view PET/CT: a head-to-head intra-individual comparison of the Biograph Vision Quadra with the Biograph Vision PET/CT. Eur J Nucl Med Mol Imaging. 2021;48:2395–404. https://doi.org/10.1007/s00259-021-05282-7.
Johnson J, Alahi A, Fei-Fei L. Perceptual losses for real-time style transfer and super-resolution. European conference on computer vision: Springer; 2016. p. 694–711.
Shan H, Zhang Y, Yang Q, Kruger U, Kalra MK, Sun L, et al. 3-D convolutional encoder-decoder network for low-dose CT via transfer learning from a 2-D trained network. IEEE Trans Med Imaging. 2018;37:1522–34.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Comput Sci. 2014.
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13:600–12.
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. Tensorflow: large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:160304467. 2016.
Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014.
Hu Z, Xue H, Zhang Q, Gao J, Zhang N, Zou S, et al. DPIR-Net: direct PET image reconstruction based on the Wasserstein generative adversarial network. IEEE Trans Radiation X Plasma Med Sci. 2021;5:35–43. https://doi.org/10.1109/trpms.2020.2995717.
Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021;3:199–217.
Daube-Witherspoon ME, Muehllehner G. Treatment of axial data in three-dimensional PET. J Nucl Med. 1987;28:1717–24.
Gundlich B, Musmann P, Weber S, Nix O, Semmler W. From 2D PET to 3D PET: issues of data representation and image reconstruction. Z Med Phys. 2006;16:31–46. https://doi.org/10.1078/0939-3889-00290.
Prenosil GA, Sari H, Fürstner M, Afshar-Oromieh A, Shi K, Rominger A, et al. Performance characteristics of the Biograph Vision Quadra PET/CT system with a long axial field of view using the NEMA NU 2–2018 standard. J Nucl Med. 2022;63:476–84. https://doi.org/10.2967/jnumed.121.261972.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. https://doi.org/10.1038/nature14539.
Webb S. Deep learning for biology. Nature. 2018;554:555–7. https://doi.org/10.1038/d41586-018-02174-z.
Towards trustable machine learning. Nat Biomed Eng. 2018;2:709–10. https://doi.org/10.1038/s41551-018-0315-x.
Wang S, Cao G, Wang Y, Liao S, Wang Q, Shi J, et al. Review and prospect: artificial intelligence in advanced medical imaging. Front Radiol. 2021;1. https://doi.org/10.3389/fradi.2021.781868.
Acknowledgements
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided.
Funding
This project received funding from China’s National Natural Science Foundation [Grant No. 11875036], Tsinghua University Initiative Scientific Research Program, and the Swiss National Science Foundation [SNSF Grant No. 188350 and 188914].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by RM, JH, HS, and SX. The first draft of the manuscript was written by RM, JH, and KS, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. WL, RQ, AR, JL, and KS supported this work with funding acquisition.
Corresponding authors
Ethics declarations
Ethics approval
The study was conducted in accordance with the requirements of the respective local ethics committees in Switzerland (Req-2021–00517).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
The authors affirm that human research participants provided informed consent for publication of Figs. 4, 5, 6, 7.
Conflict of interests
Hasan Sari is a full-time employee of Siemens Healthcare AG, Switzerland. No other potential conflict of interest relevant to this article was reported.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, R., Hu, J., Sari, H. et al. An encoder-decoder network for direct image reconstruction on sinograms of a long axial field of view PET. Eur J Nucl Med Mol Imaging 49, 4464–4477 (2022). https://doi.org/10.1007/s00259-022-05861-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-022-05861-2