Abstract
High temperature is one of the major environmental stresses affecting plant growth and fitness. Heat stress transcription factors (HSFs) play critical roles in regulating the expression of heat-responsive genes. However, how HSFs are regulated remains obscure. Here, we show that ALBA4, ALBA5 and ALBA6, which phase separate into stress granules (SGs) and processing bodies (PBs) under heat stress, directly bind selected messenger RNAs, including HSF mRNAs, and recruit them into SGs and PBs to protect them from degradation under heat stress in Arabidopsis. The alba456 triple mutants, but not single and double mutants, display pleiotropic developmental defects and hypersensitivity to heat stress. Mutations in XRN4, a cytoplasmic 5′ to 3′ exoribonuclease, can rescue the observed developmental and heat-sensitive phenotypes of alba456 seedlings. Our study reveals a new layer of regulation for HSFs whereby HSF mRNAs are stabilized by redundant action of ALBA proteins in SGs and PBs for plant thermotolerance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The eCLIP-seq sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive under the accession number PRJNA750641. The eCLIP-seq data for Pro35S:GFP at 22 °C were downloaded from GEO (GSE128619). No new code was generated in this study. The accession numbers of the genes used in this research are as follows: ALBA4 (AT1G20220), ALBA5 (AT1G76010), ALBA6 (AT3G07030), DCP5 (AT1G26110), PAB2 (AT4G34110), PAB4 (AT2G23350), Rbp47b (AT3G19130), DCP1 (AT1G08370), DCP2 (AT5G13570), EIF4A1 (AT3G13920), PAT1 (AT1G79090), XRN4/EIN5 (AT1G54490), HSP101 (AT1G74310), HSFA7B (AT3G63350), HSFA2 (AT2G26150), HSFB2A (AT5G62020), MBF1C (AT3G24500), DREB2A (AT5G05410), ACTIN2 (AT3G18780). Source data are provided with this paper.
References
Hasanuzzaman, M., Nahar, K., Alam, M. M., Roychowdhury, R. & Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684 (2013).
Ding, Y., Shi, Y. & Yang, S. Molecular regulation of plant responses to environmental temperatures. Mol. Plant 13, 544–564 (2020).
Guo, M. et al. The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7, 114 (2016).
Buchan, J. R. & Parker, R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 (2009).
Chantarachot, T. et al. DHH1/DDX6-like RNA helicases maintain ephemeral half-lives of stress-response mRNAs. Nat. Plants 6, 675–685 (2020).
Gutierrez-Beltran, E., Moschou, P. N., Smertenko, A. P. & Bozhkov, P. V. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 27, 926–943 (2015).
Weber, C., Nover, L. & Fauth, M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 56, 517–530 (2008).
Sorenson, R. & Bailey-Serres, J. Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl Acad. Sci. USA 111, 2373–2378 (2014).
Belostotsky, D. A. & Sieburth, L. E. Kill the messenger: mRNA decay and plant development. Curr. Opin. Plant Biol. 12, 96–102 (2009).
Aizer, A. et al. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J. Cell Sci. 127, 4443–4456 (2014).
Jang, G. J., Yang, J. Y., Hsieh, H. L. & Wu, S. H. Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. Proc. Natl Acad. Sci. USA 116, 6451–6456 (2019).
Decker, C. J. & Parker, R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4, a012286 (2012).
Xu, J., Yang, J. Y., Niu, Q. W. & Chua, N. H. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18, 3386–3398 (2006).
Thomas, M. G., Loschi, M., Desbats, M. A. & Boccaccio, G. L. RNA granules: the good, the bad and the ugly. Cell Signal. 23, 324–334 (2011).
Anderson, P. & Kedersha, N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430–436 (2009).
Vanderweyde, T., Youmans, K., Liu-Yesucevitz, L. & Wolozin, B. Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology 59, 524–533 (2013).
Iserman, C. et al. Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell 181, 818–831 (2020).
Laurens, N. et al. Alba shapes the archaeal genome using a delicate balance of bridging and stiffening the DNA. Nat. Commun. 3, 1328 (2012).
Zhang, N., Guo, L. & Huang, L. The Sac10b homolog from Sulfolobus islandicus is an RNA chaperone. Nucleic Acids Res. 48, 9273–9284 (2020).
Mani, J. et al. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS One 6, e22463 (2011).
Aravind, L., Iyer, L. M. & Anantharaman, V. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 4, R64 (2003).
Yuan, W. et al. ALBA protein complex reads genic R-loops to maintain genome stability in Arabidopsis. Sci. Adv. 5, eaav9040 (2019).
Thandapani, P., O’Connor, T. R., Bailey, T. L. & Richard, S. Defining the RGG/RG motif. Mol. Cell 50, 613–623 (2013).
Jang, G. J., Jang, J. C. & Wu, S. H. Dynamics and functions of stress granules and processing bodies in plants. Plants (Basel) 9, 1122 (2020).
Xu, J. & Chua, N. H. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21, 3270–3279 (2009).
Motomura, K. et al. Diffuse decapping enzyme DCP2 accumulates in DCP1 foci under heat stress in Arabidopsis thaliana. Plant Cell Physiol. 56, 107–115 (2015).
Perea-Resa, C. et al. The LSM1-7 Complex gifferentially regulates Arabidopsis tolerance to abiotic stress conditions by promoting selective mRNA decapping. Plant Cell 28, 505–520 (2016).
Yan, C., Yan, Z., Wang, Y., Yan, X. & Han, Y. Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J. Exp. Bot. 65, 5933–5944 (2014).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Oates, M. E. et al. D(2)P(2): database of disordered protein predictions. Nucleic Acids Res. 41, D508–D516 (2013).
Jones, D. T. & Cozzetto, D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857–863 (2015).
McLoughlin, F., Kim, M., Marshall, R. S., Vierstra, R. D. & Vierling, E. HSP101 interacts with the proteasome and promotes the clearance of ubiquitylated protein aggregates. Plant Physiol. 180, 1829–1847 (2019).
Zhu, D. et al. The features and gegulation of co-transcriptional splicing in Arabidopsis. Mol. Plant 13, 278–294 (2020).
Kosmacz, M. et al. Protein and metabolite composition of Arabidopsis stress granules. New Phytol. 222, 1420–1433 (2019).
Kastenmayer, J. P. & Green, P. J. Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl Acad. Sci. USA 97, 13985–13990 (2000).
Souret, F. F., Kastenmayer, J. P. & Green, P. J. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15, 173–183 (2004).
Markmiller, S. et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604 (2018).
Gutierrez-Beltran, E. et al. Tudor staphylococcal nuclease is a docking platform for stress granule components and is essential for SnRK1 activation in Arabidopsis. EMBO J. 40, e105043 (2021).
Aulas, A. et al. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 130, 927–937 (2017).
Unsworth, H., Raguz, S., Edwards, H. J., Higgins, C. F. & Yague, E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 24, 3370–3380 (2010).
Hubstenberger, A. et al. P-Body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144–157 (2017).
Khong, A. et al. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820 (2017).
Goyal, M., Banerjee, C., Nag, S. & Bandyopadhyay, U. The Alba protein family: structure and function. Biochim. Biophys. Acta 1864, 570–583 (2016).
Mateju, D. et al. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183, 1801–1812 (2020).
Verma, J. K. et al. OsAlba1, a dehydration-responsive nuclear protein of rice (Oryza sativa L. ssp. indica), participates in stress adaptation. Phytochemistry 100, 16–25 (2014).
Chene, A. et al. PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites. Nucleic Acids Res. 40, 3066–3077 (2012).
Wang, L. C. et al. Arabidopsis HIT4 encodes a novel chromocentre-localized protein involved in the heat reactivation of transcriptionally silent loci and is essential for heat tolerance in plants. J. Exp. Bot. 64, 1689–1701 (2013).
Wang, Z. P. et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 (2015).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Li, Q. et al. Regulation of active DNA demethylation by a methyl-CpG-binding domain protein in Arabidopsis thaliana. PLoS Genet. 11, e1005210 (2015).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Khan, A. & Mathelier, A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics 18, 287 (2017).
Gu, Z., Eils, R., Schlesner, M. & Ishaque, N. EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Bioinformatics 18, 287 (2017).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Tian, T. et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–w129 (2017).
Kuret, K., Amalietti, A. G. & Ule, J. Positional motif analysis reveals the extent of specificity of protein-RNA interactions observed by CLIP. Preprint at bioRxiv https://doi.org/10.1101/2021.12.07.471544 (2021).
Acknowledgements
We thank Q.-J. Chen for the CRISPR–Cas9 system. This study was supported by the National Key R&D Programme of China to W.Q. (grant no. 2018YFE0204700) and the National Natural Science Foundation of China to L.-M.F. (grant no. 32170285) and W.Q. (grant no. 31970614).
Author information
Authors and Affiliations
Contributions
J.T. and W.Q. designed the research. J.T., Z.R., S.Z., D.C., W.Y. and W.Q. performed the experiments. L.S., Y.H., W.W., Z.W. and W.Q. analysed the data. J.T., L.-M.F. and W.Q. wrote the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Chung-Mo Park, Hong-Xuan Lin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Interactions between ALBA4/ALBA5/ALBA6 and DCP5.
a, Yeast two-hybrid analysis of the interactions between ALBA proteins and PB and SG components. Combinations of constructs were transformed into yeast strain AH109 and assayed on medium lacking Leu and Trp (SD-TL), medium lacking Leu, Trp, and His (SD-TLH), and medium lacking Leu, Trp, His and Ade (SD-TLHA). AD, activating domain; BD, binding domain; Vec, empty vector. Data are representative of three independent experiments. b, Co-IP results showing the interactions between ALBA proteins and DCP5 in vivo. Transgenic plants expressing ALBA4-Myc or ALBA5-Myc and F1 offspring seedlings (ProALBA4:ALBA4-Myc × ProDCP5:DCP5-GFP and Pro35S:ALBA5-Myc × ProDCP5:DCP5-GFP) were used for Co-IP. Data are representative of two independent experiments with similar results.
Extended Data Fig. 2 ALBA4, ALBA5 and ALBA6 reversibly localize to cytoplasmic mRNP granules under heat stress.
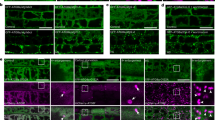
a, b, Fluorescence images showing colocalization of ALBA6 with ALBA4 (a) and ALBA5 (b). The root tissue of 5-day-old F1 seedlings co-expressing the indicated transgenes under control (22 °C) and heat stress (39 °C for 50 min) conditions was used for the experiments. Scale bars = 10 μm. Images are representative of three independent replicates, and at least five roots were monitored for each condition and replicate. c, Fluorescence images showing localization of ALBA4, ALBA5, ALBA6 and Rbp47b during recovery from heat stress. The transgenic plants were pretreated at 39 °C for 50 min, after which images of root tip cells were taken at 22 °C at the indicated time points. Scale bars = 10 μm. d, Quantification of ALBA4, ALBA5, ALBA6 and Rbp47b foci in (c). Data represent the mean ± SD. For ALBA4-GFP foci, n = 17, 11, 14, 14 and 10, for 10, 30, 50, 70 and 90 min, respectively; for ALBA5-GFP foci, n = 13, 18, 12, 17 and 8, for 10, 30, 50, 70 and 90 min, respectively; for ALBA6-GFP foci, n = 18, 23, 29, 26 and 24, for 10, 30, 50, 70 and 90 min, respectively; for Rbp47b, n = 30, 23, 25, 18 and 15, for 10, 30, 50, 70 and 90 min, respectively.
Extended Data Fig. 3 ALBA5 colocalizes with SG and PB marker proteins under heat stress.
a, b, Fluorescence images showing colocalization of ALBA5 with SG marker proteins Rbp47b (a) and PAB2 (b). c-e, Fluorescence images showing colocalization of ALBA5 with PB marker proteins DCP1 (c), DCP2 (d) and DCP5 (e). The root tissue of 5-day-old F1 seedlings co-expressing the indicated transgenes under control (22 °C) and heat stress (39 °C for 50 min) conditions was used for the experiments. Scale bars = 10 μm. Images are representative of three independent replicates, and at least five roots were monitored for each condition and replicate.
Extended Data Fig. 4 ALBAs undergo liquid-liquid phase separation in vivo and in vitro.
a, FRAP of ALBA5-GFP foci in the cytoplasm of Arabidopsis root tip cells. Time 0 indicates the time of the photobleaching pulse. Arrowheads indicate the bleached foci. Scale bars = 0.5 μm. Data are representative of four independent experiments with similar results. b, Dynamics of ALBA5-GFP foci in the cytoplasm of Arabidopsis root tip cells after heat stress treatment. Plants were pre-treated at 39 °C for 20 min, and high-resolution images were acquired at 39 °C continuously. Scale bars = 0.5 μm. Data are representative of three independent experiments with similar results. c, FRAP of ALBA6-GFP foci in the cytoplasm of Arabidopsis root tip cells. Scale bars = 0.5 μm. Data are representative of four independent experiments with similar results. d,e, Recovery curves of ALBA5-GFP (d) and ALBA6-GFP (e) cytoplasmic foci after photobleaching. Data are presented as mean ± SD (n = 4). f, Analysis of ALBA4 (16 μM) droplet formation in vitro. Scale bar = 10 μm. Data are representative of three independent experiments with similar results.
Extended Data Fig. 5 ALBA4, ALBA5 and ALBA6 act redundantly to regulate plant development.
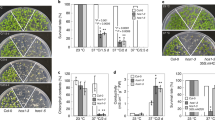
a, b, Phenotypes (a) and quantification of the fresh weight (b) of 10-day-old wild-type Col-0, single mutant (alba4, alba5 and alba6), double mutant (alba45, alba46 and alba56) and triple mutant (alba456-3) seedlings. c, d, Primary root growth phenotype (c) and quantification of the primary root length (d) of 8-day-old Col-0 and alba mutant seedlings. In (b) and (d), dark horizontal line, median; edges of boxes, 25th (bottom) and 75th (top) percentiles; whiskers, minimum and maximum. Significant differences between two genotypes are indicated by different letters (P < 0.05, one-way ANOVA with post hoc Tukey HSD tests; n = 30). See Source Data for P values.
Extended Data Fig. 6 ALBA4, ALBA5 or ALBA6 transgene can rescue the developmental phenotypes of alba456.
a, b, Representative photographs (a) and quantification of the fresh weight (b) of 10-day-old Col-0, alba456-2 and ALBA4/5/6 complementation lines. GFP-tagged ALBA4 and ALBA5 were driven by their native promoters, and GFP-tagged ALBA6 was driven by the 35 S promoter. c,d, Primary root growth phenotype (c) and quantification of the primary root length (d) of 8-day-old Col-0, alba456-2 and ALBA4/5/6 complementation lines. In (b) and (d), dark horizontal line, median; edges of boxes, 25th (bottom) and 75th (top) percentiles; whiskers, minimum and maximum. Significant differences between two genotypes are indicated by different letters (P < 0.05, one-way ANOVA with post hoc Tukey HSD tests; n = 30). See Source Data for P values.
Extended Data Fig. 7 ALBA4-GFP and DCP5-GFP transgenes can rescue the heat hypersensitive phenotype of alba456-2 and dcp5-1, respectively.
a, b, Photographs of Col-0, alba456-2, ALBA4-GFP-complemented alba456-2, hsp101, dcp5-1, DCP5-GFP-complemented dcp5-1 seedlings under control (22 °C), heat shock (44 °C for 90 min) and prolonged heat treatment (37 °C for 5 d). Plants in each treatment group were grown on the same plate. At least three independent biological replicates were conducted. Results from one representative biological replicate are shown. The hsp101 mutant was included as a control. Scale bars = 2 cm. c, Quantification of the data shown in (a) and (b). The percentage of seedlings in different phenotypic classes was determined for different genotypes and different conditions. Data are presented as the mean ± SD of three biological replicates. Significant differences between two groups are marked with different letters as determined by two-sided Fisher’s exact test (P < 0.01). See Source Data for P values.
Extended Data Fig. 8 ALBA5 binds ssRNA in vitro and in vivo.
a, Coomassie blue staining of His-tagged ALBA5 purified from E.coli. Data are representative of five independent experiments with similar results. b, EMSA results showing ALBA5 binding to ssRNA probes and competition by unlabeled ssRNA probes. 3′-Fam-labeled substrates (80 nM) were incubated with increasing concentrations of ALBA5. The protein concentrations in lanes 1, 2, 3 and 4 were 0, 1, 2 and 3 μM, respectively. For competition analysis, 32 μM of unlabeled substrate was used as a cold probe and incubated with 3 μM of ALBA5-His and 80 nM of 3′-Fam-labeled substrates. For EMSA, at least three independent biological replicates were performed, and representative results are shown. c, Principal component analysis (PCA) plots of ALBA5 binding clusters in 5-day-old seedlings under control (22 °C) and heat stress (39 °C for 50 min) conditions.
Extended Data Fig. 9 Mutations in XRN4 can rescue the heat hypersensitivity of alba456-2.
a, Schematic presentation of the Arabidopsis XRN4 gene. Untranslated and coding regions are depicted as thick gray and blue boxes, respectively. Introns are depicted as thin black boxes. Triangles indicate the CRISPR/Cas9 target sites and T-DNA insertion position in XRN4. The XRN4 gene is drawn to scale. The sgRNA target sequences are highlighted in yellow. The PAM sites are underlined. b, Photographs of Col-0, alba456-2, xrn4-5 and alba456-2 xrn4-9 seedlings under control (22 °C) and prolonged heat treatment (37 °C for 5 d). Scale bars = 2 cm. c, Quantification of the data shown in (b); the percentage of seedlings in different phenotypic classes was determined for different genotypes and different conditions. Data are presented as the mean ± SD of three biological replicates. Significant differences between two groups are marked with different letters as determined by two-sided Fisher’s exact test (P < 0.01). d, Decay profiles of HSF mRNAs under heat stress conditions (37 °C for 12 h). ALBA5 non-target AT2G07689 was used as the negative control. The mRNA decay profile of each genotype is presented as the relative mRNA abundance after transcription inhibition by cordycepin. Results from one representative replicate of two independent biological replicates are shown (each in three technical replicates). Data represent mean ± SD. See Source Data for P values.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8.
Supplementary Data 1
Identification of co-purified proteins of ALBA4, ALBA5, ALBA6, DCP5 and Rb47b by mass spectrometry.
Supplementary Data 2
List of ALBA5 binding sites from eCLIP-seq.
Supplementary Data 3
Primers, sgRNAs and probes used in this study.
Supplementary Data 4
Statistical source data for Supplementary Figures.
Source data
Source Data for Fig. 3
Statistical source data for Fig.3.
Source Data for Fig. 4
Statistical source data for Fig.4.
Source Data for Fig. 5
Uncropped scans of gels for Fig. 5g.
Source Data for Fig, 6
Statistical source data for Fig.6.
Source Data for Fig. 7
Statistical source data for Fig.7.
Source Data for Extended Data Fig. 1
Uncropped western blots for Extended Data Fig. 1b.
Source Data for Extended Data Fig. 5
Statistical source data for Extended Data Fig.5.
Source Data for Extended Data Fig. 6
Statistical source data for Extended Data Fig.6.
Source Data for Extended Data Fig. 7
Statistical source data for Extended Data Fig.7.
Source Data for Extended Data Fig. 9
Statistical source data for Extended Data Fig.9.
Rights and permissions
About this article
Cite this article
Tong, J., Ren, Z., Sun, L. et al. ALBA proteins confer thermotolerance through stabilizing HSF messenger RNAs in cytoplasmic granules. Nat. Plants 8, 778–791 (2022). https://doi.org/10.1038/s41477-022-01175-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-022-01175-1
This article is cited by
-
The processing body component VARICOSE plays a multiplayer role towards stress management in Arabidopsis
Plant Physiology Reports (2024)