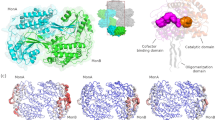
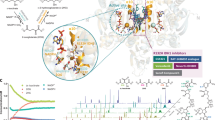
Abstract
Aldehyde dehydrogenases (ALDHs) are promising cancer drug targets, as certain isoforms are required for the survival of stem-like tumor cells. We have discovered selective inhibitors of ALDH1B1, a mitochondrial enzyme that promotes colorectal and pancreatic cancer. We describe bicyclic imidazoliums and guanidines that target the ALDH1B1 active site with comparable molecular interactions and potencies. Both pharmacophores abrogate ALDH1B1 function in cells; however, the guanidines circumvent an off-target mitochondrial toxicity exhibited by the imidazoliums. Our lead isoform-selective guanidinyl antagonists of ALDHs exhibit proteome-wide target specificity, and they selectively block the growth of colon cancer spheroids and organoids. Finally, we have used genetic and chemical perturbations to elucidate the ALDH1B1-dependent transcriptome, which includes genes that regulate mitochondrial metabolism and ribosomal function. Our findings support an essential role for ALDH1B1 in colorectal cancer, provide molecular probes for studying ALDH1B1 functions and yield leads for developing ALDH1B1-targeting therapies.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Any data generated or analyzed during this study, associated protocols and materials are available from the corresponding author on reasonable request. X-ray crystal structures have been validated and deposited with the Protein Data Bank with the following entries: 7MJC (ALDH1B1–NAD+), 7MJD (ALDH1B1–NAD+–2) and 7RAD (ALDH1B1–NAD+–IGUANA-4). Raw and processed proteomics data for the TPP study are publicly available in the MassIVE repository (MassIVE ID MSv000088824), which is a member of the ProteomeXchange Consortium (ProteomeXchange ID PXD031630). Raw and processed RNA-seq data for the identification of ALDH1B1-dependent genes are publicly available in the Gene Expression Omnibus database (GEO accession number GSE165621). Source data are provided with this paper.
References
Vassalli, G. Aldehyde dehydrogenases: not just markers, but functional regulators of stem cells. Stem Cells Int. 2019, 3904645 (2019).
Xu, X. et al. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett. 369, 50–57 (2015).
Marcato, P. et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells 29, 32–45 (2011).
Mao, P. et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl Acad. Sci. USA 110, 8644–8649 (2013).
Luo, Y. et al. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells 30, 2100–2113 (2012).
Shao, C. et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin. Cancer Res. 20, 4154–4166 (2014).
Landen, C. N. Jr et al. Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer. Mol. Cancer Ther. 9, 3186–3199 (2010).
Singh, S. et al. ALDH1B1 is crucial for colon tumorigenesis by modulating Wnt/β-catenin, Notch and PI3K/Akt signaling pathways. PLoS ONE 10, e0121648 (2015).
Mameishvili, E. et al. Aldh1b1 expression defines progenitor cells in the adult pancreas and is required for Kras-induced pancreatic cancer. Proc. Natl Acad. Sci. USA 116, 20679–20688 (2019).
Meng, E. et al. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS ONE 9, e107142 (2014).
Li, B., Yang, K., Liang, D., Jiang, C. & Ma, Z. Discovery and development of selective aldehyde dehydrogenase 1A1 (ALDH1A1) inhibitors. Eur. J. Med. Chem. 209, 112940 (2021).
Gelardi, E. L. M. et al. A selective competitive inhibitor of aldehyde dehydrogenase 1A3 hinders cancer cell growth, invasiveness and stemness in vitro. Cancers 13, 356 (2021).
Li, J. et al. A specific inhibitor of ALDH1A3 regulates retinoic acid biosynthesis in glioma stem cells. Commun. Biol. 4, 1420 (2021).
Stagos, D. et al. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab. Dispos. 38, 1679–1687 (2010).
Chen, Y. et al. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem. Biophys. Res. Commun. 405, 173–179 (2011).
Singh, S. et al. ALDH1B1 links alcohol consumption and diabetes. Biochem. Biophys. Res. Commun. 463, 768–773 (2015).
Singh, S. et al. Aldehyde dehydrogenase 1B1 as a modulator of pancreatic adenocarcinoma. Pancreas 45, 117–122 (2016).
Hom, M. E., Ondrus, A. E., Sakata-Kato, T., Rack, P. G. & Chen, J. K. Bicyclic imidazolium inhibitors of Gli transcription factor activity. ChemMedChem 15, 1044–1049 (2020).
Luo, M. & Tanner, J. J. Structural basis of substrate recognition by aldehyde dehydrogenase 7A1. Biochemistry 54, 5513–5522 (2015).
Riveros-Rosas, H., Gonzalez-Segura, L., Julian-Sanchez, A., Diaz-Sanchez, A. G. & Munoz-Clares, R. A. Structural determinants of substrate specificity in aldehyde dehydrogenases. Chem. Biol. Interact. 202, 51–61 (2013).
Sarvi, S. et al. ALDH1 bio-activates nifuroxazide to eradicate ALDHHigh melanoma-initiating cells. Cell Chem. Biol. 25, 1456–1469 (2018).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Tang, Z. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017).
Putzbach, W. et al. Many si/shRNAs can kill cancer cells by targeting multiple survival genes through an off-target mechanism. eLife 6, e29702 (2017).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Chen, X. et al. LGR5 is required for the maintenance of spheroid-derived colon cancer stem cells. Int. J. Mol. Med. 34, 35–42 (2014).
Reily, C. et al. Mitochondrially targeted compounds and their impact on cellular bioenergetics. Redox Biol. 1, 86–93 (2013).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Franken, H. et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 10, 1567–1593 (2015).
Vodenkova, S. et al. 5-Fluorouracil and other fluoropyrimidines in colorectal cancer: past, present and future. Pharmacol. Ther. 206, 107447 (2020).
Chou, T. C. Drug combination studies and their synergy quantification using the Chou–Talalay method. Cancer Res. 70, 440–446 (2010).
Neal, J. T. et al. Organoid modeling of the tumor immune microenvironment. Cell 175, 1972–1988 (2018).
Lo, Y. H. et al. A CRISPR/Cas9-engineered ARID1A-deficient human gastric cancer organoid model reveals essential and nonessential modes of oncogenic transformation. Cancer Discov. 11, 1562–1581 (2021).
Giroux, V. et al. Mouse intestinal Krt15+ crypt cells are radio-resistant and tumor initiating. Stem Cell Rep. 10, 1947–1958 (2018).
Nakanishi, Y. et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat. Genet. 45, 98–103 (2013).
Yang, L. et al. Pregnancy-specific glycoprotein 9 (PSG9), a driver for colorectal cancer, enhances angiogenesis via activation of SMAD4. Oncotarget 7, 61562–61574 (2016).
Casazza, A. et al. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell 24, 695–709 (2013).
De Vlaeminck, Y. et al. Targeting neuropilin-1 with nanobodies reduces colorectal carcinoma development. Cancers 12, 3582 (2020).
Cox, P. J., Phillips, B. J. & Thomas, P. The enzymatic basis of the selective action of cyclophosphamide. Cancer Res. 35, 3755–3761 (1975).
Weiner, H., Hu, J. H. & Sanny, C. G. Rate-limiting steps for the esterase and dehydrogenase reaction catalyzed by horse liver aldehyde dehydrogenase. J. Biol. Chem. 251, 3853–3855 (1976).
Morgan, C. A. & Hurley, T. D. Characterization of two distinct structural classes of selective aldehyde dehydrogenase 1A1 inhibitors. J. Med. Chem. 58, 1964–1975 (2015).
Yang, S. M. et al. Discovery of orally bioavailable, quinoline-based aldehyde dehydrogenase 1A1 (ALDH1A1) inhibitors with potent cellular activity. J. Med. Chem. 61, 4883–4903 (2018).
Huddle, B. C. et al. Structure-based optimization of a novel class of aldehyde dehydrogenase 1A (ALDH1A) subfamily-selective inhibitors as potential adjuncts to ovarian cancer chemotherapy. J. Med. Chem. 61, 8754–8773 (2018).
Snyder, V., Reed-Newman, T. C., Arnold, L., Thomas, S. M. & Anant, S. Cancer stem cell metabolism and potential therapeutic targets. Front. Oncol. 8, 203 (2018).
Bastide, A. & David, A. The ribosome, (slow) beating heart of cancer (stem) cell. Oncogenesis 7, 34 (2018).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020).
Cho, Y. H. et al. 5-FU promotes stemness of colorectal cancer via p53-mediated WNT/β-catenin pathway activation. Nat. Commun. 11, 5321 (2020).
Cong, J. et al. A novel chemoradiation targeting stem and nonstem pancreatic cancer cells by repurposing disulfiram. Cancer Lett. 409, 9–19 (2017).
Taipale, J. et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406, 1005–1009 (2000).
Hoppel, C., DiMarco, J. P. & Tandler, B. Riboflavin and rat hepatic cell structure and function. Mitochondrial oxidative metabolism in deficiency states. J. Biol. Chem. 254, 4164–4170 (1979).
Wolfram, S. et al. A small azide-modified thiazole-based reporter molecule for fluorescence and mass spectrometric detection. Beilstein J. Org. Chem. 10, 2470–2479 (2014).
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Chen, C. H., Cruz, L. A. & Mochly-Rosen, D. Pharmacological recruitment of aldehyde dehydrogenase 3A1 (ALDH3A1) to assist ALDH2 in acetaldehyde and ethanol metabolism in vivo. Proc. Natl Acad. Sci. USA 112, 3074–3079 (2015).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
DeLano, W. L. Pymol: an open-source molecular graphics tool. CPP4 Newsletter Pro. Crystallogr. 40, 82–92 (2002).
Campeau, E. et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE 4, e6529 (2009).
Jafari, R. et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 9, 2100–2122 (2014).
Chen, J. K., Taipale, J., Young, K. E., Maiti, T. & Beachy, P. A. Small molecule modulation of Smoothened activity. Proc. Natl Acad. Sci. USA 99, 14071–14076 (2002).
Tran, U., Zhang, G. C., Eom, R., Billingsley, K. L. & Ondrus, A. E. Small molecule intervention in a protein kinase C-Gli transcription factor axis. ACS Chem. Biol. 15, 1321–1327 (2020).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B. Stat. Methodol. 57, 289–300 (1995).
Acknowledgements
We thank T. Hurley for the vector for bacterial ALDH1B1 expression, E. Patton for ALDH1A3–/– A375 cells, P. Beachy for Shh-LIGHT2 cells, G. Ponhert for BOPIDY azide and S. Swick for assistance with compound characterization. This work was supported by the National Institutes of Health (R35 GM127030, R01 CA244334 and R01 GM113100 to J.K.C.; U01 CA217851 and U54 CA224081 to C.J.K.; R01 AA11147 to D.M.-R. and F32 CA183527 to A.E.O.), SPARK at Stanford (J.K.C.), Weston Havens Foundation (J.K.C.), the Emerson Collective (J.K.C. and C.J.K.), the Human Cancer Models Initiative (C.J.K.) and Stand Up to Cancer (C.J.K.). Flow cytometry experiments were performed at the Stanford Shared FACS Facility supported by an NIH S10 Shared Instrument grant (S10 OD026831). Use of the Stanford University Mass Spectrometry facility is supported, in part, by the National Institutes of Health (P30 CA124435) utilizing the Stanford Cancer Institute Proteomics/Mass Spectrometry Shared Resources. Use of the SSRL/SLAC National Accelerator Laboratory is supported by the U.S. Department of Energy, Office of Science and Office of Basic Energy Sciences, under contract number DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health (P41 GM103393). NMR experiments included spectra acquired on a Bruker Avance NEO 500 MHz spectrometer supported by an NIH S10 Shared Instrument Grant (S10 OD028697).
Author information
Authors and Affiliations
Contributions
Z.F., M.E.H., T.E.B., Z.C.R., D.F., A.E.O., A.K.M., M.G.T., C.J.K. and J.K.C. designed the experiments and analyzed the data. A.E.O., T.K. and J.K.C. designed compounds and synthetic routes, and A.E.O. prepared compounds. A.E.O. conducted photoaffinity cross-linking, click chemistry, two-dimensional gel electrophoresis and mass spectrometry analysis, and C.-H.C. assisted with the two-dimensional gel electrophoresis. Z.F., M.E.H., T.E.B., Z.C.R. and C.-H.C. expressed and purified ALDH proteins. Z.F., M.E.H., T.E.B., Z.C.R. and C.R.M. performed the enzyme kinetics assays and compound activity profiling. Z.F., Z.C.R. and D.F. conducted the X-ray crystallography studies of ALDH1B1 and ALDH1B1–inhibitor complexes. Z.F. and C.R.M. performed computational modeling of ALDH1–inhibitor complexes. Z.F., T.E.B. and Z.C.R. conducted spheroid and adherent culture growth assays. Z.F., M.E.H. and T.E.B. performed the ALDEFLUOR assays. Z.F. and M.E.H. conducted the Seahorse, HEK-293T viability and Shh-LIGHT2 signaling assays. T.E.B. prepared the ALDH1B1 mutants. Z.F. and Y.G. generated the ALDH1B1–/– and ALDH1B1 rescue clones, and Z.F. conducted related growth assays. Z.F. generated the 5-FU-resistant SW480 cells and conducted related growth assays. A.K.M. and M.G.T. conducted the organoid experiments. Z.F. and Y.G. prepared total RNA samples for RNA-seq analysis, and Z.F. and J.K.C. analyzed the transcriptomic data. Z.F. conducted CETSAs and prepared TPP samples. T.E.B analyzed the TPP data. Z.F. and J.K.C. wrote the manuscript. A.E.O., D.M.-R., C.J.K. and J.K.C. acquired funding for the project.
Corresponding author
Ethics declarations
Competing interests
J.K.C., Z.F., M.E.H., T.K., C.R.M. and A.E.O. have filed a PCT application related to the compounds described in this study.
Peer review
Peer review information
Nature Chemical Biology thanks Ronald Buckanovich, John Tanner and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Photoaffinity labeling of imidazolium-binding proteins.
a, Photoaffinity labeling and TAMRA-azide tagging of live NIH-3T3 cells, demonstrating the mitochondrial localization of the imidazolium target. Scale bar: 10 μm. b, Mouse liver mitochondria were photocrosslinked with probe 3 in the absence or presence of competitor 2. The mitochondria were then homogenized, reacted with BODIPY azide (no competitior) or MegaStrokes 673 azide (competitor), and resolved by two-dimensional gel electrophoresis. Protein spots that were specifically labeled by 3 (dashed box, spots 1-5) were then isolated for mass spectrometry-based sequencing.
Extended Data Fig. 2 Inhibition of cellular ALDH1B1 activity by imidazolium derivatives.
a, Immunofluorescence staining of ALDH1A3–/– A375 cells transiently transfected with ALDH1B1, demonstrating the mitochondrial localization of the exogenous protein. Scale bar: 40 µm. b, Flow cytometry-based assays of ALDH1B1 activity and its pharmacological inhibition using ALDH1A3–/– A375 cells. The cells were transiently transfected with ALDH1B1 cDNA or a vector control, incubated with the designated compounds, and then treated with ALDEFLUOR reagent. The cells were gated by ALDEFLUOR signal intensity and side scatter area (SSC-A) to identify those with ALDH1B1 activity, and the percentage of cells outside of the negative control gate is shown for each condition. DMSO and the pan-ALDH inhibitor DEAB were used as negative and positive controls, respectively. c, Chemical structure of 63, which is inactive against ALDH1B1. d, Activity of 63 against selected ALDH isoforms. Fold-change values are relative to a DMSO control and are the average of three biological replicates ± s.d. e, Fluorescence-activated cell sorting (FACS) plots of ALDH1B1-overexpressing ALDH1A3–/– A375 cells that were incubated with DMSO or 63 and then treated with ALDEFLUOR reagent.
Extended Data Fig. 3 ALDH1B1 promotes SW480 and HCT116 cell growth in spheroid culture.
a,e, Western blot detection of ALDH1B1 protein in individual SW480 and HCT116 cell clones that were transiently transfected with Cas9 cDNA and ALDH1B1 gRNA-1 and gRNA-2. Lysates from the parental lines (P) are also shown, and importin β1 (KPNB1) was used as a loading control. SW480 clone 3 and clone 2 were used as ALDH1B1+/+ and ALDH1B1–/– clones for subsequent studies. b,f, Phase-contrast micrographs of spheroid cultures derived from SW480 and HCT116 cells with differing ALDH1B1 genotypes. c,g, Quantification of spheroid sizes for the micrographs shown in b and f. Each dot represents an individual spheroid with an area that is >500 µm2 in the images. Error bars represent the average spheroid size ± s.e.m. d,h, Viability of the ALDH1B1–/– clone in either adherent or spheroid conditions, as determined by cellular ATP levels and normalized to that of the as ALDH1B1+/+ clone. Data are the average of four (d and h, adherent), eight (d, spheroid) and six (h, spheroid) biological replicates ± s.e.m. Scale bars: 1 mm.
Extended Data Fig. 4 Exogenous ALDH1B1 rescues the spheroid growth of ALDH1B1 knockout colon cancer cells.
a, Western blot analysis of SW480 clones with the indicated ALDH1B1 genotypes and lentivirally transduced with either EGFP or exogenous ALDH1B1. Importin β1 (KNPB1) was used as gel-loading control. b, Spheroid cultures of the SW480 clones described in a. c, Quantification of spheroid sizes for the micrographs shown in b. Each dot represents an individual spheroid with an area that is >500 µm2 in the image. Error bars represent the average spheroid size ± s.e.m. d, Viability of the ALDH1B1–/– SW480 clone transduced with EGFP or ALDH1B1 and cultured in either adherent or spheroid conditions, as determined by cellular ATP levels and normalized to that of the ALDH1B1+/+ clone transduced with EGFP. Data are the average of four biological replicates ± s.e.m.
Extended Data Fig. 5 Molecular basis of ALDH1B1-IGUANA binding.
a,b, Lineweaver Burk plots demonstrating that IGUANA-4 exhibits non-competitive inhibition with respect to acetaldehyde (a) and uncompetitive inhibition with respect to NAD+ (b). Both enzyme kinetics assays used the same series of inhibitor concentrations, and the data are the average of three biological replicates ± s.e.m. c, Wall-eyed stereoview of the ALDH1B1–NAD+/2 (light green cartoon and green stick model) and ALDH1B1–NAD+/–IGUANA-4 (light blue cartoon and blue stick model) complexes shown as superimposed structures. Residues 143–154 and 490–500 and the NAD+ cofactor were omitted for clarity. d, Wall-eyed stereoview of the electron density map for IGUANA-4 (blue mesh) bound to ALDH1B1. The polder omit map was calculated with coefficients mFo-DFc and is contoured at 4σ. Residues in the inhibitor-binding site are shown, and the red dashed line indicates a potential n-to-π* interaction between the Asn457 backbone carbonyl and the guanidine (3.2 Å).
Extended Data Fig. 6 IGUANAs engage ALDH1B1 in live cells.
a, Flow cytometry-based ALDEFLUOR assays using ALDH1A3–/– A375 cells, demonstrating the ability of IGUANA-3 to inhibit cellular ALDH1B1 activity. The cells were transiently transfected with ALDH1B1 cDNA or a vector control, incubated with IGUANA-3 or DMSO vehicle alone, and then treated with ALDEFLUOR reagent. The cells were gated by ALDEFLUOR signal intensity and side scatter area (SSC-A) to identify those with ALDH1B1 activity, and the percentage of cells outside of the negative control gate is shown for each condition. b, Cellular thermal shift assay demonstrating that IGUANA-1 stabilizes endogenous ALDH1B1 in live SW480 cells. Western blot signals for ALDH1B1 and total protein staining of the soluble fraction are shown for each condition. c, Corresponding melting curves of endogenous ALDH1B1 in the presence and absence of IGUANA-1. Data are the average of two biological replicates ± s.d., normalized to the DMSO condition at 45 °C.
Extended Data Fig. 7 IGUANAs suppress colon cancer spheroid growth.
a, Brightfield micrographs of SW480 spheroid cultures treated with IGUANA-3 and then stained with crystal violet. b, Quantification of spheroid sizes for the micrographs shown in b. Each dot represents an individual spheroid with an area that is >500 µm2 in the micrograph. Error bars represent the average spheroid size ± s.e.m. c Brightfield, fluorescent, and merged micrographs of SW480 spheroids treated with IGUANA-1 or DMSO vehicle alone for 3 days and then stained overnight with the viability dye SYTOX Green. Scale bars: a, 1 mm; c, 100 µm.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Figs. 1–21, Note (synthetic procedures and compound characterization) and full-length blots for Supplementary figures.
Supplementary Data 1
Chemical structures of imidazolium and guanidine derivatives.
Supplementary Data 2
Mass spectrometry data for photoaffinity-labeled protein spots.
Supplementary Data 3
TPP data.
Supplementary Data 4
RNA-seq data for the ALDH1B1-dependent transcriptome.
Supplementary Data 5
Statistical source data for Supplementary figures.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed blots.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
About this article
Cite this article
Feng, Z., Hom, M.E., Bearrood, T.E. et al. Targeting colorectal cancer with small-molecule inhibitors of ALDH1B1. Nat Chem Biol 18, 1065–1075 (2022). https://doi.org/10.1038/s41589-022-01048-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01048-w
This article is cited by
-
Interrogating colorectal cancer metastasis to liver: a search for clinically viable compounds and mechanistic insights in colorectal cancer Patient Derived Organoids
Journal of Experimental & Clinical Cancer Research (2023)
-
Targeting ALDH1B1 in colorectal cancer
Nature Reviews Drug Discovery (2022)