Abstract
In the past decade, the application of machine learning (ML) to healthcare has helped drive the automation of physician tasks as well as enhancements in clinical capabilities and access to care. This progress has emphasized that, from model development to model deployment, data play central roles. In this Review, we provide a data-centric view of the innovations and challenges that are defining ML for healthcare. We discuss deep generative models and federated learning as strategies to augment datasets for improved model performance, as well as the use of the more recent transformer models for handling larger datasets and enhancing the modelling of clinical text. We also discuss data-focused problems in the deployment of ML, emphasizing the need to efficiently deliver data to ML models for timely clinical predictions and to account for natural data shifts that can deteriorate model performance.
Similar content being viewed by others
Main
In the past decade, machine learning (ML) for healthcare has been marked by particularly rapid progress. Initial groundwork has been laid for many healthcare needs that promise to improve patient care, reduce healthcare workload, streamline healthcare processes and empower the individual1. In particular, ML for healthcare has been successful in the translation of computer vision through the development of image-based triage2 and second readers3. There has also been rapid progress in the harnessing of electronic health records4,5 (EHRs) to predict the risk and progression of many diseases6,7. A number of software platforms for ML are beginning to make their way into the clinic8. In 2018, iDX-DR, which detects diabetic retinopathy, was the first ML system for healthcare that the United States Food and Drug Administration approved for clinical use8. Babylon9, a chatbot triage system, has partnered with the United Kingdom’s National Healthcare system. Furthermore, Viz.ai10,11 has rolled out their triage technology to more than 100 hospitals in the United States.
As ML systems begin to be deployed in clinical settings, the defining challenge of ML in healthcare has shifted from model development to model deployment. In bridging the gap between the two, another trend has emerged: the importance of data. We posit that large, well-designed, well-labelled, diverse and multi-institutional datasets drive performance in real-world settings far more than model optimization12,13,14, and that these datasets are critical for mitigating racial and socioeconomic biases15. We realize that such rich datasets are difficult to obtain, owing to clinical limitations of data availability, patient privacy and the heterogeneity of institutional data frameworks. Similarly, as ML healthcare systems are deployed, the greatest challenges in implementation arise from problems with the data: how to efficiently deliver data to the model to facilitate workflow integration and make timely clinical predictions? Furthermore, once implemented, how can model robustness be maintained in the face of the inevitability of natural changes in physician and patient behaviours? In fact, the shift from model development to deployment is also marked by a shift in focus: from models to data.
In this Review, we build on previous surveys1,16,17 and take a data-centric approach to reviewing recent innovations in ML for healthcare. We first discuss deep generative models and federated learning as strategies for creating larger and enhanced datasets. We also examine the more recent transformer models for handling larger datasets. We end by highlighting the challenges of deployment, in particular, how to process and deliver usable raw data to models, and how data shifts can affect the performance of deployed models.
Deep generative models
Generative adversarial networks (GANs) are among the most exciting innovations in deep learning in the past decade. They offer the capability to create large amounts of synthetic yet realistic data. In healthcare, GANs have been used to augment datasets18, alleviate the problems of privacy-restricted19 and unbalanced datasets20, and perform image-modality-to-image-modality translation21 and image reconstruction22 (Fig. 1). GANs aim to model and sample from the implicit density function of the input data23. They consist of two networks that are trained in an adversarial process under which one network, the ‘generator’, generates synthetic data while the other network, the ‘discriminator’, discriminates between real and synthetic data. The generative model aims to implicitly learn the data distribution from a set of samples to further generate new samples drawn from the learned distribution, while the discriminator pushes the generator network to sample from a distribution that more closely mirrors the true data distribution.
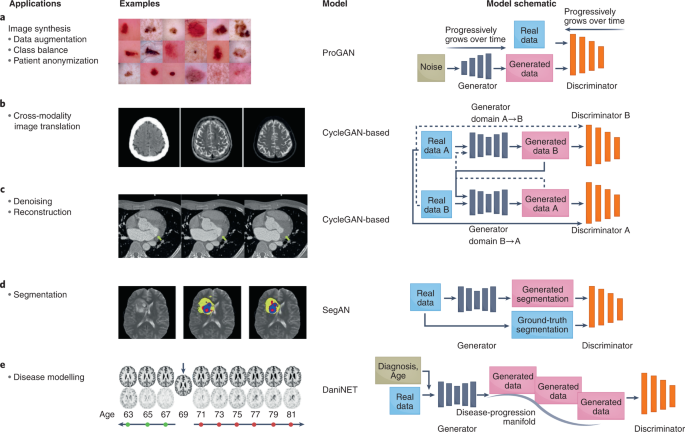
a, GANs can be used to augment datasets to increase model performance and anonymize patient data. For example, they have been used to generate synthetic images of benign and malignant lesions from real images183. b, GANs for translating images acquired with one imaging modality into another modality51. Left to right: input CT image, generated MR image and reference MR image. c, GANs for the denoising and reconstruction of medical images184. Left, low-dose CT image of a patient with mitral valve prolapse, serving as the input into the GAN. Right, corresponding routine-dose CT image and the target of the GAN. Middle, GAN-generated denoised image resembling that obtained from routine-dose CT imaging. The yellow arrows indicate a region that is distinct between the input image (left) and the target denoised image (right). d, GANs for image classification, segmentation and detection39. Left, input image of T2 MRI slice from the multimodal brain-tumour image-segmentation benchmark dataset. Middle, ground-truth segmentation of the brain tumour. Right, GAN-generated segmentation image. Yellow, segmented tumour; blue, tumour core; and red, Gd-enhanced tumour core. e, GANs can model a spectrum of clinical scenarios and predict disease progression66. Top: given an input MR image (denoted by the arrow), DaniGAN can generate images that reflect neurodegeneration over time. Bottom, difference between the generated image and the input image. ProGAN, progressive growing of generative adversarial network; DaniNet, degenerative adversarial neuroimage net. Credit: Images (‘Examples’) reproduced with permission from: a, ref. 183, Springer Nature Ltd; b, ref. 51, under a Creative Commons licence CC BY 4.0; c, ref. 184, Wiley; d, ref. 39, Springer Nature Ltd; e, ref. 66, Springer Nature Ltd.
Over the years, a multitude of GANs have been developed to overcome the limitations of the original GAN (Table 1), and to optimize its performance and extend its functionalities. The original GAN23 suffered from unstable training and low image diversity and quality24. In fact, training two adversarial models is, in practice, a delicate and often difficult task. The goal of training is to achieve a Nash equilibrium between the generator and the discriminator networks. However, simultaneously obtaining such an equilibrium for networks that are inherently adversarial is difficult and, if achieved, the equilibrium can be unstable (that is, it can be suddenly lost after model convergence). This has also led to sensitivity to hyperparameters (making the tuning of hyperparameters a precarious endeavour) and to mode collapse, which occurs when the generator produces a limited and repeated number of outputs. To remedy these limitations, changes have been made to GAN architectures and loss functions. In particular, the deep convolutional GAN (DCGAN25), a popular GAN often used for medical-imaging tasks, aimed to combat instability by introducing key architecture-design decisions, including the replacement of fully connected layers with convolutional layers, and the introduction of batch normalization (to standardize the inputs to a layer when training deep neural networks) and ReLU (rectified linear unit) activation. The Laplacian pyramid of adversarial networks GAN (LAPGAN26) and the progressively growing GAN (ProGAN27) build on DCGAN to improve training stability and image quality. Both LAPGAN and ProGAN start with a small image, which promotes training stability, and progressively grow the image into a higher-resolution image.
The conditional GAN (cGAN28) and the auxiliary classifier GAN (AC-GAN29) belong to a subtype of GANs that enable the model to be conditioned on external information to create synthetic data of a specific class or condition. This was found to improve the quality of the generated samples and increase the capability to handle the generation of multimodal data. The pix2pix GAN30, which is conditioned on images, allows for image-to-image translation (also across imaging modalities) and has been popular in healthcare applications.
A recent major architectural change to GANs involve attention mechanisms. Attention was first introduced to facilitate language translation and has rapidly become a staple in deep-learning models, as it can efficiently capture longer-range global and spatial relations from input data. The incorporation of attention into GANs has led to the development of self-attention GANs (SAGANs)31,32 and BigGAN;33; the latter scales up SAGAN to achieve state-of-the-art performance.
Another primary strategy to mitigate the limitations of GANs involves improving the loss function. Early GANs used the Jensen-Shannon divergence and the Kullback-Leibler divergence as loss functions to minimize the difference in distribution between the synthetic generated dataset and the real-data dataset. However, the Jensen-Shannon divergence was found to fail in scenarios where there is no overlap (or little overlap) between distributions, while the minimization of the Kullback-Leibler divergence can lead to mode collapse. To address these problems, a number of GANs have used alternative loss functions. The most popular are arguably the Wasserstein GAN (WGAN34) and the Wasserstein GAN gradient penalty (WGAN-GP35). The Wasserstein distance measures the effort to minimize the distance between dataset distributions and has been shown to have a smoother gradient. Additional popular strategies that have been implemented to improve GAN performance and that do not involve modifying the model architecture include spectral normalization and varying how frequently the discriminator is updated (with respect to the update frequency of the generator).
The explosive progress of GANs has spawned many more offshoots of the original GAN, as documented by the diverse models that now populate the GAN Model Zoo36.
Augmenting datasets
In the past decade, many deep-learning models for medical-image classification3,37, segmentation38,39 and detection40 have achieved physician-level performance. However, the success of these models is ultimately beholden to large, diverse, balanced and well-labelled datasets. This is a bottleneck that extends across domains, yet it is particularly restrictive in healthcare applications where collecting comprehensive datasets comes with unique obstacles. In particular, large amounts of standardized clinical data are difficult to obtain, and this is exacerbated by the reality that clinical data often reflects the patient population of one or few institutions (with the data sometimes overrepresenting common diseases or healthy populations and making the sampling of rarer conditions more difficult). Datasets with high class imbalance or insufficient variability can often lead to poor model performance, generalization failures, unintentional modelling of confounders41 and propagation of biases42. To mitigate these problems, clinical datasets can be augmented by using standard data-manipulation techniques, such as the flipping, rotation, scaling and translation of images43. However, these methods can lead to limited increases in performance and generate highly correlated training data.
GANs offer potent solutions to these problems. GANs can be used to augment training data to improve model performance. For example, a convolutional neural network (CNN) for the classification of liver lesions, trained on both synthetically and traditionally augmented data, boosted the performance of the model by 10% with respect to a CNN trained on only traditionally augmented datasets18. Moreover, when generating synthetic data across data classes, developing a generator for each class can result in higher model performance20,44, as was shown via the comparison of two variants of GANs (a DCGAN that generated labelled examples for each of three lesion classes separately and an AC-GAN that incorporated class conditioning to generate labelled examples)18.
The aforementioned studies involved class-balanced datasets but did not address medical data with either simulated or real class imbalances. In an assessment of the capability of GANs to alleviate the shortcomings of unbalanced chest-X-ray datasets20, it was found that training a classifier on real unbalanced datasets that had been augmented with DCGANs outperformed models that were trained with the unbalanced and balanced versions of the original dataset. Although there was an increase in classification accuracy across all classes, the greatest increase in performance was seen in the most imbalanced classes (pneumothorax and oedema), which had just one-fourth the number of training cases as the next class.
Protecting patient privacy
The protection of patient privacy is often a leading concern when developing clinical datasets45. Sharing patient data when generating multi-institution clinical datasets can pose a risk to patient privacy46. Even if privacy protocols are followed, patient characteristics can sometimes be inferred from the ML model and its outputs47,48. In this regard, GANs may provide a solution. Data created by GANs cannot be attributed to a single patient, as they synthesize data that reflect the patient population in aggregate. GANs have thus been used as a patient-anonymization tool to generate synthetic data for model training9,49. Although models trained on just synthetic data can perform poorly, models trained on synthetic data and fine-tuned with 10% real data resulted in similar performance to models trained on real datasets augmented with synthetic data19. Similarly, using synthetic data generated from GANs to train an image-segmentation model was sufficient to achieve 95% of the accuracy of the same model trained on real data49. Hence, using synthetic data during model development can mitigate potential patient-privacy violations.
Image-to-image translation
One exciting use of GANs involves image-to-image translation. In healthcare, this capability has been used to translate between imaging modalities—between computed tomography (CT) and magnetic resonance (MR) images21,49,50,51, between CT and positron emission tomography (PET)52,53,54, between MR and PET55,56,57, and between T1 and T2 MR images58,59. Transfer between image modalities can reduce the need for additional costly and time-intensive image acquisitions, can be used in scenarios where imaging is not possible (as is the case for MR imaging in individuals with metal implants) and to expand the types of training data that can be created from image datasets. There are two predominant strategies for image-to-image translation: paired-image training (with pix2pix30) and unpaired training (with CycleGAN60). For example, pix2pix was used to generate synthetic CT images for accurate MR-based dose calculations for the pelvis61. Similarly, using paired magnetic resonance angiography and MR images, pix2pix was modified to generate a model for the translation of T1 and T2 MR images to retrospectively inspect vascular structures62.
Obtaining paired images can be difficult in scenarios involving moving organs or multimodal medical images that are in three dimensions and do not have cross-modality paired data. In such cases, one can use CycleGAN60, which handles image-to-image translation on unpaired images. A difficulty with unpaired images is the lack of ground-truth labels for evaluating the accuracy of the predictions (yet real cardiac MR images have been used to compare the performance of segmentation models trained on synthetic cardiac MR images translated from CT images49). Another common problem is the need to avoid geometric distortions that destroy anatomical structures. Limitations with geometric distortions can be overcome by using two auxiliary mappings to constrain the geometric invariance of synthetic data21.
Opportunities
In the context of clinical datasets, GANs have primarily been used to augment or balance the datasets, and to preserve patient privacy. Yet a burgeoning application of GANs is their use to systematically explore the entire terrain of clinical scenarios and disease presentations. Indeed, GANs can be used to generate synthetic data to combat model deterioration in the face of domain shifts63,64, for example, by creating synthetic data that simulate variable lighting or camera distortions, or that imitate data collected from devices from different vendors or from different imaging modalities. Additionally, GANs can be used to create data that simulate the full spectrum of clinical scenarios and disease presentations, from dangerous and rare clinical scenarios such as incorrect surgery techniques63, to modelling the spectrum of brain-tumour presentation19, to exploring the disease progression of neurodegenerative diseases65,66.
However, GANs can suffer from training instability and low image diversity and quality. These limitations could hamper the deployment of GANs in clinical practice. For example, one hope for image-to-image translation in healthcare involves the creation of multimodality clinical images (from CT and MR, for example) for scenarios in which only one imaging modality is possible. However, GANs are currently limited in the size and quality of the images that they can produce. This raises the question of whether these images can realistically be used clinically when medical images are typically generated at high resolution. Moreover, there may be regulatory hurdles involved in approving ML healthcare models that have been trained on synthetic data. This is further complicated by the current inability to robustly evaluate and control the quality of GANs and of the synthetic data that they generate67. Still, in domains unrelated to healthcare, GANs have been used to make tangible improvements to deployed models68. These successes may lay a foundation for the real-world application of GANs in healthcare.
Federated learning
When using multi-institutional datasets, model training is typically performed centrally: data siloed in individual institutions are aggregated into a single server. However, data used in such ‘centralized training’ represent a fraction of the vast amount of clinical data that could be harnessed for model development. Yet, openly sharing and exchanging patient data is restricted by many legal, ethical and administrative constraints; in fact, in many jurisdictions, patient data must remain local.
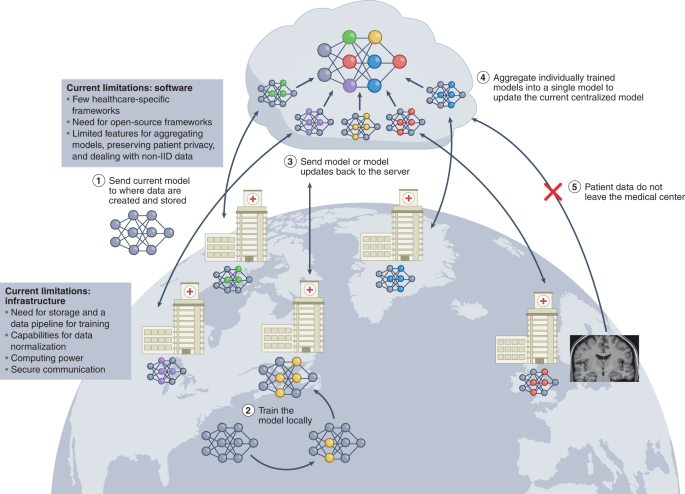
Federated learning is a paradigm for training ML models when decentralized data are used collaboratively under the orchestration of a central server69,70 (Fig. 2). In contrast to centralized training, where data from various locations are moved to a single server to train the model, federated learning allows for the data to remain in place. At the start of each round of training, the current copy of the model is sent to each location where the training data are stored. Each copy of the model is then trained and updated using the data at each location. The updated models are then sent from each location back to the central server, where they are aggregated into a global model. The subsequent round of training follows, the newly updated global model is distributed again, and the process is repeated until model convergence or training is stopped. At no point do the data leave a particular location or institution, and only individuals associated with an institution have direct access to its data. This mitigates concerns about privacy breaches, minimizes costs associated with data aggregation, and allows training datasets to quickly scale in size and diversity. The successful implementation of federated learning could transform how deep-learning models for healthcare are trained. Here we focus on two applications: cross-silo federated learning and cross-device federated learning (Table 2).
Multiple institutions collaboratively train an ML model. Federated learning begins when each institution notifies a central server of their intention to participate in the current round of training. Upon notification, approval and recognition of the institution, the central server sends the current version of the model to the institution (step 1). Then, the institution trains the model locally using the data available to it (step 2). Upon completion of local training, the institution sends the model back to the central server (step 3). The central server aggregates all of the models that have been trained locally by each of the individual institutions into a single updated model (step 4). This process is repeated in each round of training until model training concludes. At no point during any of the training rounds do patient data leave the institution (step 5). The successful implementation of federated learning requires healthcare-specific federated learning frameworks that facilitate training, as well as institutional infrastructure for communication with the central server and for locally training the model.
Cross-silo federated learning
Cross-silo federated learning is an increasingly attractive solution to the shortcomings of centralized training71. It has been used to leverage EHRs to train models to predict hospitalization due to heart disease72, to promote the development of ‘digital twins’ or ‘Google for patients’73, and to develop a Coronavirus disease 2019 (COVID-19) chest-CT lesion segmenter74. Recent efforts have focused on empirically evaluating model-design parameters, and on logistical decisions to optimize model performance and overcome the unique implementation challenges of federated learning, such as bottlenecks in protecting privacy and in tackling the statistical heterogeneity of the data75,76.
Compared with centralized training, one concern of federated learning is that models may encounter more severe domain shifts or overfitting. However, models trained through federated learning were found to achieve 99% of the performance of traditional centralized training even with imbalanced datasets or with relatively few samples per institution, thus showing that federated learning can be realistically implemented without sacrificing performance or generalization77,78.
Although federated learning offers greater privacy protection because patient data are no longer being transmitted, there are risks of privacy breaches79. Communicating model updates during the training process can reveal sensitive information to third parties or to the central server. In certain instances, data leakage can occur, such as when ML models ‘memorize’ datasets80,81,82 and when access to model parameters and updates can be used to infer the original dataset83. Differential privacy84 can further reinforce privacy protection for federated learning70,85,86. Selective parameter sharing87 and the sparse vector technique88 are two strategies for achieving greater privacy, but at the expense of model performance (this is consistent with differential-privacy findings in domains outside of medicine and healthcare80,89).
Another active area of research for federated learning in healthcare involves the handling of data that are neither independent nor identically distributed (non-IID data). Healthcare data are particularly susceptible to this problem, owing to a higher prevalence of certain diseases in certain institutions (which can cause label-distribution skew) or to institution-specific data-collection techniques (leading to ‘same label, different features’ or to ‘same features, different label’). Many federated learning strategies assume IID data, but non-IID data can pose a very real problem in federated learning; for example, it can cause the popular federated learning algorithm FedAvg70 to fail to converge90. The predominant strategies for addressing this issue have involved the reframing of the data to achieve a uniform distribution (consensus solutions) or the embracing of the heterogeneity of the data69,91,92 (pluralistic solutions). In healthcare, the focus has been on consensus solutions involving data sharing (a small subset of training data is shared among all institutions93,94).
Cross-device federated learning to handle health data from individuals
‘Smart’ devices can produce troves of continuous, passive and individualized health data that can be leveraged to train ML models and deliver personalized health insights for each user1,16,39,95,96. As smart devices become increasingly widespread, and as computing and sensor technology become more advanced and cheaper to mass-produce, the amount of health data will grow exponentially. This will accentuate the challenges of aggregating large quantities of data into a single location for centralized training and exacerbate privacy concerns (such as any access to detailed individual health data by large corporations or governments).
Cross-device federated learning was developed to address the increasing amounts of data that are being generated ‘at the edge’ (that is, by decentralized smart devices), and has been deployed on millions of smart devices; for example, for voice recognition (by Apple, for the voice assistant Siri97) and to improve query suggestions (by Google, for the Android operating system98).
The application of cross-device federated learning to train healthcare models for smart devices is an emerging area of research. For example, using a human-activity-recognition dataset, a global model (FedHealth) was pre-trained using 80% of the data before deploying it to be locally trained and then aggregated99. The aggregated model was then sent back to each user and fine-tuned on user-specific data to develop a personalized model for the user. Model personalization resolves issues arising from the highly different probability distributions that may arise across users and the global model. This training strategy outperformed non-federated learning by nearly 5.3%.
Limitations and opportunities
In view of the initial promises and successes of federated learning, the next few years will be defined by progress towards the implementation of federated learning in healthcare. This will require a high degree of coordination across institutions at each step of the federated learning process. Before training, medical data will need to undergo data normalization and standardization. This can be challenging, owing to differences in how data are collected, stored, labelled and partitioned across institutions. Current data pre-processing pipelines could be adapted to create multi-institutional training datasets, yet in federated learning, the responsibility shifts from a central entity to each institution individually. Hence, methods to streamline and validate these processes across institutions will be essential for the successful implementation of federated learning.
Another problem concerns the inability of the developer of the model to directly inspect data during model development. Data inspection is critical for troubleshooting and for identifying any mislabelled data as well as general trends. Tools (such as Federated Analytics, developed by Google100) that use GANs to create synthetic data that resemble the original training data101 and derive population-level summary statistics from the data, can be helpful. However, it is currently unclear whether tools that have been developed for cross-device settings can be applied to cross-silo healthcare settings while preserving institutional privacy.
Furthermore, federated learning will require robust frameworks for the implementation of federated networks. Many such software is proprietary, and many of the open-source frameworks are primarily intended for use in research. The primary concerns of federated learning can be addressed by frameworks designed to reinforce patient privacy, facilitate model aggregation and tackle the challenges of non-IID data.
One main hurdle is the need for each participating healthcare institution to acquire the necessary infrastructure. This implies ensuring that each institution has the same federated learning framework and version, that stable and encrypted network communication is available to send and receive model updates from the central server, and that the computing capabilities (institutional graphics processing units or access to cloud computing) are sufficient to train the model. Although most large healthcare institutions may have the necessary infrastructure in place, it has typically been optimized to store and handle data centrally. The adaptation of infrastructure to handle the requirements of federated learning requires coordinated effort and time.
A number of ongoing federated learning initiatives in healthcare are underway. Specifically, the Federated Tumour Segmentation Initiative (a collaboration between Intel and the University of Pennsylvania) trains lesion-segmentation models collaboratively across 29 international healthcare institutions102. This initiative focuses on finding the optimal algorithm for model aggregation, as well as on ways to standardize training data from various institutions. In another initiative (a collaboration of NVIDIA and several institutions), federated learning was used to train mammography-classification models103. These efforts may establish blueprints for coordinated federated networks applied to healthcare.
Natural language processing
Harnessing natural language processing (NLP)—the automated understanding of text—has been a long-standing goal for ML in healthcare1,16,17. NLP has enabled the automated translation of doctor–patient interactions to notes5,104,105, the summarization of clinical notes106, the captioning of medical images107,108 and the prediction of disease progression6,7. However, the inability to efficiently train models using the large datasets needed to achieve adept natural-language understanding has limited progress. In this section, we provide an overview of two recent innovations that have transformed NLP: transformers and transfer learning for NLP. We also discuss their applications in healthcare.
Transformers
When modelling sequential data, recurrent neural networks (RNNs) have been the predominant choice of neural network. In particular, long short-term memory networks109 and gated units110 were staple RNNs in modelling EHR data, as these networks can model the sequential nature of clinical data111,112 and clinical text5,104,105,113. However, RNNs harbour several limitations114. Namely, RNNs process data sequentially and not in parallel. This restricts the size of the input datasets and of the networks, which limits the complexity of the features and the range of relations that can be learned114. Hence, RNNs are difficult to train, deploy and scale, and are suboptimal for capturing long-range patterns and global patterns in data. However, learning global or long-range relationships are often needed when learning language representations. For example, sentences far removed from a word may be important for providing context for the word, and previous clinical events that have occurred can inform clinical decisions that are made years later. For a period, CNNs, which are adept at parallelization, were used to overcome some of the limitations of RNNs115, but were found to be inefficient when modelling longer global dependencies.
In 2017, a research team at Google (the Google Brain team) released the transformer, a landmark model that has revolutionized NLP116. Compared with RNN and CNN models, transformers are more parallelizable and less computationally complex at each layer, and thus can handle larger training data and learn longer-range and global relations. The use of only attention layers for the encoders and decoders while forgoing the use of RNNs or CNNs was critical to the success of transformers. Attention was introduced and refined117,118 to handle bottlenecks in sequence-to-sequence RNNs110,119. Attention modules allow models to globally relate different positions of a sequence to compute a richer representation of the sequence116, and does so in parallel, allowing for increased computing efficiency and for the embedding of longer relations of the input sequence (Fig. 3).
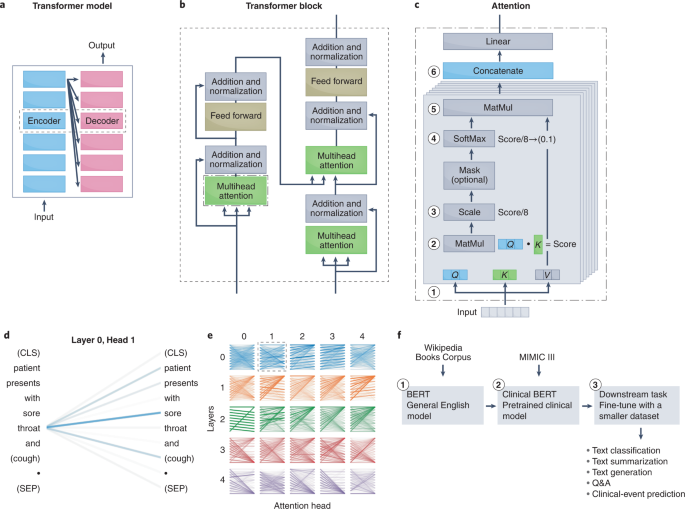
a, The original transformer model performs language translation, and contains encoders that convert the input into an embedding and decoders that convert the embedding into the output. b, The transformer model uses attention mechanisms within its encoders and decoders. The attention module is used in three places: in the encoder (for the input sentence), in the decoder (for the output sentence) and in the encoder–decoder in the decoder (for embeddings passed from the encoder). c, The key component of the transformer block is the attention module. Briefly, attention is a mechanism to determine how much weight to place on input features when creating embeddings for downstream tasks. For NLP, this involves determining how much importance to place on surrounding text when creating a representation for a particular word. To learn the weights, the attention mechanism assigns a score to each pair of words from an input phrase to determine how strongly the words should influence the representation. To obtain the score, the transformer model first decomposes the input into three vectors: the query vector (Q; the word of interest), the key vector (K; surrounding words) and the value vector (V; the contents of the input) (1). Next, the dot product is taken between the query and key vector (2) and then scaled to stabilize training (3). The SoftMax function is then applied to normalize the scores and ensure that they add to 1 (4). The output SoftMax score is then multiplied by the value vector to apply a weighted focus to the input (5). The transformer model has multiple attention mechanisms (termed attention heads); each learn a separate representation for the same word, which therefore increases the relations that can be learned. Each attention head is composed of stacked attention layers. The output of each attention mechanism is concatenated into a single matrix (6) that is fed into the downstream feed-forward layer. d,e, Visual representation of what is learned185. Lines relate the query (left) to the words that are attended to the most (right). Line thickness denotes the magnitude of attention, and colours represent the attention head. d, The learned attention in one attention-mechanism layer of one head. e, Examples of what is learned by each layer of each attention head. Certain layers learn to attend to the next words (head 2, layer 0) or to the previous word (head 0, layer 0). f, Workflow for applying a transformer language model to a clinical task. Matmul, matrix multiplication; (CLS), classification token placed at the start of a sentence to store the sentence-level embedding; (SEP), separation token placed at the end of a sentence. BERT, bidirectional encoder representations from transformers; MIMIC, multiparameter intelligence monitoring in intensive care.
Transfer learning for NLP
Simultaneous and subsequent work following the release of the transformer resolved another main problem in NLP: the formalization of the process of transfer learning. Transfer learning has been used most extensively in computer vision, owing to the success of the ImageNet challenge, which made pre-trained CNNs widely available120. Transfer learning has enabled the broader application of deep learning in healthcare17, as researchers can fine-tune a pre-trained CNN adept at image classification on a smaller clinical dataset to accomplish a wide spectrum of healthcare tasks3,37,121,122. Until recently, robust transfer learning for NLP models was not possible, which limited the use of NLP models in domain-specific applications. A series of recent milestones have enabled transfer learning for NLP. The identification of the ideal pre-training language task for deep-learning NLP models (for example, masked-language modelling, predicting missing words from surrounding context, next-sentence prediction or predicting whether two sentences follow one another) was solved by universal language model fine-tuning (ULM-FiT123) and embeddings from language model (ELMo124). The generative pre-trained transformer (GPT125) from Open AI and the bidirectional encoder representations from transformers (BERT126) from Google Brain then applied the methods formalized by ULM-FiT and ELMo to transformer models, delivering pre-trained models that achieved unprecedented capabilities on a series of NLP tasks.
Transformers for the understanding of clinical text
Following the success of transformers for NLP, their potential to handle domain-specific text, specifically clinical text, was quickly assessed. The performances of the transformer-based model BERT, the RNN-based model ELMo and traditional word-vector embeddings127,128 at clinical-concept extraction (the identification of the medical problems, tests and treatments) from EHR data were evaluated106. BERT outperformed traditional word vectors by a substantial margin and was more computationally efficient than ELMo (it achieved higher performance with fewer training iterations)129,130,131,132. Pre-training on a dataset of 2 million clinical notes (the dataset multiparameter intelligence monitoring in intensive care132; MIMIC-III) increased the performance of all NLP models. This suggests that contextual embeddings encode valuable semantic information not accounted for in traditional word representations106. However, the performance of MIMIC-III BERT began to decline after achieving its optimal model; this is perhaps indicative of the model losing information learned from the large open corpus and converging to a model similar to the one initialized from scratch106. Hence, there may be a fine balance between learning from a large open-domain corpus and a domain-specific clinical corpus. This may be a critical consideration when applying pre-trained models to healthcare tasks.
To facilitate the further application of clinically pre-trained BERT129 to downstream clinical tasks, a BERT pre-trained on large clinical datasets was publicly released. Because transformers and deep NLP models are resource-intensive to train (training the BERT model can cost US$50,000–200,000133; and pre-training BERT on clinical datasets required 18 d of continuous training, an endeavour that may be out of the reach of many institutions), openly releasing pre-trained clinical models can facilitate widespread advancements of NLP tasks in healthcare. Other large and publicly available clinically pre-trained models (Table 3) are ClinicalBERT130, BioBERT134 and SciBERT135.
The release of clinically pre-trained models has spurred downstream clinical applications. ClinicalBERT, a BERT model pre-trained on MIMIC-III data using masked-language modelling and next-sentence prediction, was evaluated on the downstream task of predicting 30 d readmission130. Compared with previous models136,137, ClinicalBERT can dynamically predict readmission risk during a patient’s stay and uses clinical text rather than structured data (such as laboratory values, or codes from the international classification of diseases). This shows the power of transformers to unlock clinical text, a comparatively underused data source in EHRs. Similarly, clinical text from EHRs has been harnessed using SciBERT for the automated extraction of symptoms from COVID-19-positive and COVID-19-negative patients to identify the most discerning clinical presentation138. ClinicalBERT has also been adapted to extract anginal symptoms from EHRs139. Others have used enhanced clinical-text understanding for the automatic labelling and summarization of clinical reports. BioBERT and ClinicalBERT have been harnessed to extract labels from radiology text reports, enabling an automatic clinical summarization tool and labeller140. Transformers have also been used to improve clinical questioning and answering141, in clinical voice assistants142,143, in chatbots for patient triage144,145, and in medical-image-to-text translation and medical-image captioning146.
Transformers for the modelling of clinical events
In view of their adeptness to model the sequential nature of clinical text, transformers have also been harnessed to model the sequential nature of clinical events147,148,149,150,151. A key challenge of modelling clinical events is properly capturing long-term dependencies—that is, previous clinical procedures that may preclude future downstream interventions. Transformers are particularly adept at exploring longer-range relationships and were recently used to develop BEHRT152, which leverages the parallels between sequences in natural language and clinical events in EHRs to portray diagnoses as words, visits as sentences and a patient’s medical history as a document152. When used to predict the likelihood of 301 conditions in future visits, BEHRT achieved an 8–13.2% improvement over the existing state-of-the-art EHR model152. BEHRT was also used to predict the incidence of heart failure from EHR data153.
Data-limiting factors in the deployment of ML
The past decade of research in ML in healthcare has focused on model development, and the next decade will be defined by model deployment into clinical settings42,45,46,154,155. In this section, we discuss two data-centric obstacles in model deployment: how to efficiently deliver raw clinical data (Table 4) to models, and how to monitor and correct for natural data shifts that deteriorate model performance.
Delivering data to models
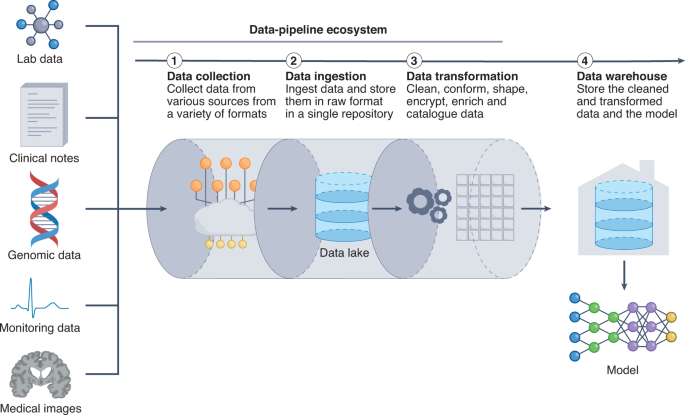
A main obstacle to model deployment is associated with how to efficiently transform raw, unstructured and heterogeneous clinical data into structured data that can be inputted into ML models. During model development, pre-processed structured data are directly inputted into the model. However, during deployment, minimizing the delay between the acquisition of raw data and the delivery of structured inputs requires an adept data pipeline for collecting data from their source, and for ingesting, preparing and transforming the data (Fig. 4). An ideal system would need to be high-throughput, have low latency and be scalable to a large number of data sources. A lack of optimization can result in major sources of inefficiency and delayed predictions from the model. In what follows, we detail the challenges of building a pipeline for clinical data and give an overview of the key components of such a pipeline.
Delivering data to a model is a key bottleneck in obtaining timely and efficient inferences. ML models require input data that are organized, standardized and normalized, often in tabular format. Therefore, it is critical to establish a pipeline for organizing and storing heterogeneous clinical data. The data pipeline involves collecting, ingesting and transforming clinical data from an assortment of data sources. Data can be housed in data lakes, in data warehouses or in both. Data lakes are central repositories to store all forms of data, raw and processed, without any predetermined organizational structure. Data in data lakes can exist as a mix of binary data (for example, images), structured data, semi-structured data (such as tabular data) and unstructured data (for example, documents). By contrast, data warehouses store cleaned, enriched, transformed and structured data with a predetermined organizational structure.
The fundamental challenge of creating an adept data pipeline arises from the need to anticipate the heterogeneity of the data. ML models often require a set of specific clinical inputs (for example, blood pressure and heart rate), which are extracted from a suite of dynamically changing health data. However, it is difficult to extract the relevant data inputs. Clinical data vary in volume and velocity (the rate that data are generated), thus prompting the question of how frequently data should be collected. Furthermore, clinical data can vary in veracity (data quality), thus requiring different pre-processing steps. Moreover, the majority of clinical data exist in an unstructured format that is further complicated by the availability of hundreds of EHR products, each with its own clinical terminology, technical specifications and capabilities156. Therefore, how to precisely extract data from a spectrum of unstructured EHR frameworks becomes critical.
Data heterogeneity must be carefully accounted for when designing the data pipeline, as it can influence throughput, latency and other performance factors. The data pipeline starts with the process of data ingestion (by which raw clinical data are moved from the data source and into the pipeline), a primary bottleneck in the throughput of the data through the pipeline. In particular, handling peaks of data generation may require the design and implementation of scalable ways to support a variable number of connected objects157. Such data-elasticity issues can take advantage of software frameworks that scale up or down in real time to more effectively use computer resources in cloud data centres158.
After the data enters the pipeline, the data-preparation stage involves the cleansing, denoising, standardization and shaping of the data into structured data that are ready for consumption by the ML system. In studies that developed data pipelines to handle healthcare data156,159,160, the data-preparation stage was found to regulate the latency of the data pipeline, as latency depended on the efficiency of the data queue, the streaming of the data and the database for storing the computation results.
A final consideration is how data should move throughout the data pipeline; specifically, whether data should move in discrete batches or in continuous streams. Batch processing involves collecting and moving source data periodically, whereas stream processing involves sourcing, moving and processing data as soon as they are created. Batch processing has the advantages of being high-throughput, comprehensive and economical (and hence may be advantageous for scalability), whereas stream processing occurs in real time (and thus may be required for time-sensitive predictions). Many healthcare systems use a combination of batch processing and stream processing160.
Established data pipelines are being harnessed to support real-time healthcare modelling. In particular, Columbia University Medical Center, in collaboration with IBM, is streaming physiological data from patients with brain injuries to predict adverse neurological complications up to 48 h before existing methods can161. Similarly, Yale School of Medicine has used a data pipeline to support real-time data acquisition for predicting the number of beds available, handling care for inpatients and patients in the intensive care unit (such as managing ventilator capacity) and tracking the number of healthcare providers exposed to COVID-19 161. However, optimizing the components of the data pipeline, particularly for numerous concurrent ML healthcare systems, remains a challenging task.
Deployment in the face of data shifts
A main obstacle in deploying ML systems for healthcare has been maintaining model robustness when faced with data shifts162. Data shifts occur when differences or changes in healthcare practices or in patient behaviour cause the deployment data to differ substantially from the training data, resulting in the distribution of the deployment data diverging from the distribution of the training data. This can lead to a decline in model performance. Also, failure to correct for data shifts can lead to the perpetuation of algorithmic biases, missing critical diagnoses163 and unnecessary clinical interventions164.
In healthcare, data shifts are common occurrences and exist primarily along the axes of institutional differences (such as local clinical practices, or different instruments and data-collection workflows), epidemiological shifts, temporal shifts (for example, changes in physician and patient behaviours over time) and differences in patient demographics (such as race, gender and age). A recent case study165 characterizing data shifts caused by institutional differences reported that pneumothorax classifiers trained on individual institutional datasets declined in performance when evaluated on data from external institutions. Similar phenomena have been observed in a number of studies41,163,166. Institutional differences are among the most patent causes of data shifts because they frequently harbour underlying differences in patient demographics, disease incidence and data-collection workflows. For example, in an analysis of chest-X-ray classifiers and their potential to generalize to other institutions, it was found that one institution collected chest X-rays using portable radiographs, whereas another used stationary radiographs41. This led to differences in disease prevalence (33% vs 2% for pneumonia) and patient demographics (average age of 63 vs 45), as portable radiographs were primarily used for inpatients who were too sick to be transported, whereas stationary radiographs were used primarily in outpatient settings. Similarly, another study found that different image-acquisition and image-processing techniques caused the deterioration of the performance of breast-mammography classifiers to random performance (areas under the receiver operating characteristic curve of 0.4–0.6) when evaluated on datasets from four external institutions and countries163. However, it is important to note that the models evaluated were trained on data collected during the 1990s and were externally tested on datasets created in 2014–2017. The decline in performance owing to temporal shifts is particularly relevant; if deployed today, models that have been trained on older datasets would be making inferences on newly generated data.
Studies that have characterized temporal shifts have provided insights into the conditions under which deployed ML models should be re-evaluated. An evaluation of models that used data collected over a period of 9 years found that model performance deteriorated substantially, drifting towards overprediction as early as one year after model development167. For the MIMIC-III dataset132 (commonly used for the development of models to predict clinical outcomes), an assessment of the effects of temporal shifts on model performance over time showed that, whereas all models experienced a moderate decline over time, the most significant drop in performance occurred owing to a shift in clinical practice, when EHRs transitioned systems164 (from CareVue to MetaVision). A modern-day analogy would be how ML systems for COVID-19 (ref. 168) that were trained on data169 acquired during the early phase of the pandemic and before the availability of COVID-19 vaccines would perform when deployed in the face of shifts in disease incidence and presentation.
Data shifts and model deterioration can also occur when models are deployed on patients with gender, racial or socioeconomic backgrounds that are different from those of the patient population that the model was trained on. In fact, it has been shown that ML models can be biased against individuals of certain races170 or genders42, or particular religious171 or socioeconomic15 backgrounds. For example, a large-scale algorithm used in many health institutions to identify patients for complex health needs underpredicted the health needs of African American patients and failed to triage them for necessary care172. Using non-representative or non-inclusive training datasets can constitute an additional source of gender, racial or socioeconomic biases. Popular chest-X-ray datasets used to train classifiers have been shown to be heavily unbalanced15: 67.6% of the patients in these datasets are Caucasian and only 8.98% are under Medicare insurance. Unsurprisingly, the performance of models trained with these datasets deteriorates for non-Caucasian subgroups, and especially for Medicare patients15. Similarly, skin-lesion classifiers that were trained primarily on images of one skin tone decrease in performance when evaluated on images of different skin tones173; in this case, the drop in performance could be attributed to variations in disease presentation that are not captured when certain patient populations are not adequately represented in the training dataset174.
These findings exemplify two underlying limitations of ML models: the models can propagate existing healthcare biases on a large scale, and insufficient diversity in the training datasets can lead to an inadequate generalization of model outputs to different patient populations. Training models on multi-institutional datasets can be most effective at combating model deterioration15, and directly combating existing biases in the training data can also mitigate their impact171. There are also solutions for addressing data shifts that involve proactively addressing them during model development175,176,177,178 or retroactively by surveilling for data shifts during model deployment179. A proactive attitude towards recognizing and addressing potential biases and data shifts will remain imperative.
Outlook
Substantial progress in the past decade has laid a foundation of knowledge for the application of ML to healthcare. In pursuing the deployment of ML models, it is clear that success is dictated by how data are collected, organized, protected, moved and audited. In this Review, we have highlighted methods that can address these challenges. The emphasis will eventually shift to how to build the tools, infrastructure and regulations needed to efficiently deploy innovations in ML in clinical settings. A central challenge will be the implementation and translation of these advances into healthcare in the face of their current limitations: for instance, GANs applied to medical images are currently limited by image resolution and image diversity, and can be challenging to train and scale; federated learning promises to alleviate problems associated with small single-institution datasets, yet it requires robust frameworks and infrastructure; and large language models trained on large public datasets can subsume racial and ethnic biases171.
Another central consideration is how to handle the regulatory assessment of ML models for healthcare applications. Current regulation and approval processes are being adapted to meet the emerging needs; in particular, initiatives are attempting to address data shifts and patient representation in the training datasets165,180,181. However, GANs, federated learning and transformer models add complexities to the regulatory process. Few healthcare-specific benchmarking datasets exist to evaluate the performance of these ML systems during clinical deployment. Moreover, the assessment of the performance of GANs is hampered by the lack of efficient and robust metrics to evaluate, compare and control the quality of synthetic data.
Notwithstanding the challenges, the fact that analogous ML technologies are being used daily by millions of individuals in other domains, most prominently in smartphones100, search engines182 and self-driving vehicles68, suggests that the challenges of deployment and regulation of ML for healthcare can also be addressed.
References
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Gulshan, V. et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316, 2402–2410 (2016).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Rajkomar, A. et al. Scalable and accurate deep learning with electronic health records. npj Digit. Med. 1, 18 (2018).
Rajkomar, A. et al. Automatically charting symptoms from patient-physician conversations using machine learning. JAMA Intern. Med. 179, 836–838 (2019).
Henry, K. E., Hager, D. N., Pronovost, P. J. & Saria, S. A targeted real-time early warning score (TREWScore) for septic shock. Sci. Transl. Med. 7, 299ra122 (2015).
Komorowski, M., Celi, L. A., Badawi, O., Gordon, A. C. & Faisal, A. A. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nat. Med. 24, 1716–1720 (2018).
Abràmoff, M. D., Lavin, P. T., Birch, M., Shah, N. & Folk, J. C. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. npj Digit. Med. 1, 39 (2018).
Iacobucci, G. Babylon Health holds talks with ‘significant’ number of NHS trusts. Brit. Med. J. 368, m266 (2020).
Hale, C. Medtronic to distribute Viz.ai’s stroke-spotting AI imaging software. Fierce Biotech (23 July 2019); https://www.fiercebiotech.com/medtech/medtronic-to-distribute-viz-ai-s-stroke-spotting-ai-imaging-software
Hassan, A. E. et al. Early experience utilizing artificial intelligence shows significant reduction in transfer times and length of stay in a hub and spoke model. Interv. Neuroradiol. 26, 615–622 (2020).
Ting, D. S. W. et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 318, 2211–2223 (2017).
McKinney, S. M. et al. International evaluation of an AI system for breast cancer screening. Nature 577, 89–94 (2020).
Liu, Y. et al. A deep learning system for differential diagnosis of skin diseases. Nat. Med. 26, 900–908 (2020).
Seyyed-Kalantari, L., Liu, G., McDermott, M., Chen, I. Y. & Ghassemi, M. CheXclusion: fairness gaps in deep chest X-ray classifiers. Pac. Symp. Biocomput. 26, 232–243 (2021).
Yu, K.-H., Beam, A. L. & Kohane, I. S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2, 719–731 (2018).
Esteva, A. et al. A guide to deep learning in healthcare. Nat. Med. 25, 24–29 (2019).
Frid-Adar, M. et al. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 321, 321–331 (2018).
Shin, H.-C. et al. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In Simulation and Synthesis in Medical Imaging SASHIMI 2018 (eds Gooya, A., Goksel, O., Oguz, I. & Burgos, N.) 1–11 (Springer Cham, 2018).
Salehinejad, H., Valaee, S., Dowdell, T., Colak, E. & Barfett, J. Generalization of deep neural networks for chest pathology classification in X-rays using generative adversarial networks. In IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 990–994 (ieeexplore.ieee.org, 2018).
Zhang, Z., Yang, L. & Zheng, Y. Translating and segmenting multimodal medical volumes with cycle-and shape-consistency generative adversarial network. In Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition 9242–9251 (IEEE, 2018).
Xu, F., Zhang, J., Shi, Y., Kang, K. & Yang, S. A fast low-rank matrix factorization method for dynamic magnetic resonance imaging restoration. In 5th International Conference on Big Data Computing and Communications (BIGCOM) 38–42 (2019).
Goodfellow, I. J. et al. Generative adversarial networks. In Advances in Neural Information Processing Systems 27 (eds .Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N. & Weinbergerm, K.Q.) Paper 1384 (Curran, 2014).
Wang, Z., She, Q. & Ward, T. E. Generative adversarial networks in computer vision: a survey and taxonomy. ACM Comput. Surv. 54, 1–38 (2021).
Radford, A., Metz, L. & Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. Preprint at https://arxiv.org/abs/1511.06434v2 (2016).
Denton, E. L., Chintala, S. & Fergus, R. Deep generative image models using a Laplacian pyramid of adversarial networks. In Advances in Neural Information Processing Systems 28 (eds Cortes, C., Lawrence, N., Lee, D., Sugiyama, M. & Garnett, R.) Paper 903 (Curran, 2015).
Karras, T., Aila, T., Laine, S. & Lehtinen, J. Progressive growing of GANs for improved quality, stability, and variation. In International Conference on Learning Representations 2018 Paper 447 (ICLR, 2018).
Mirza, M. & Osindero, S. Conditional generative adversarial nets. Preprint at https://arxiv.org/abs/1411.1784v1 (2014).
Odena, A., Olah, C. & Shlens, J. Conditional image synthesis with auxiliary classifier GANs. In Proceedings of the 34th International Conference on Machine Learning (eds. Precup, D. & Teh, Y. W.) 2642–2651 (PMLR, 2017).
Isola, P., Zhu, J.-Y., Zhou, T. & Efros, A. A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 5967–5976 (2018).
Zhang, H., Goodfellow, I., Metaxas, D. & Odena, A. Self-attention generative adversarial networks. In Proceedings of the 36th International Conference on Machine Learning (eds. Chaudhuri, K. & Salakhutdinov, R.) 7354–7363 (PMLR, 2019).
Wu, Y., Ma, Y., Liu, J., Du, J. & Xing, L. Self-attention convolutional neural network for improved MR image reconstruction. Inf. Sci. 490, 317–328 (2019).
Brock, A., Donahue, J. & Simonyan, K. Large scale GAN training for high fidelity natural image synthesis. In International Conference on Learning Representations Paper 564 (ICLR, 2019).
Arjovsky, M., Chintala, S. & Bottou, L. Wasserstein generative adversarial networks. In Proceedings of the 34th International Conference on Machine Learning (eds. Precup, D. & Teh, Y. W.) 214–223 (PMLR, 2017).
Gulrajani, I., Ahmed, F., Arjovsky, M., Dumoulin, V. & Courville, A. C. Improved training of Wasserstein GANs. In Advances in Neural Information Processing Systems 30 (eds. Guyon, I. et al.) Paper 2945 (Curran, 2017).
Hindupur, A. The-gan-zoo. https://github.com/hindupuravinash/the-gan-zoo (2018).
Rajpurkar, P. et al. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 15, e1002686 (2018).
Ouyang, D. et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 580, 252–256 (2020).
Xue, Y., Xu, T., Zhang, H., Long, L. R. & Huang, X. SegAN: adversarial network with multi-scale L1 loss for medical image segmentation. Neuroinformatics 16, 383–392 (2018).
Haque, A., Milstein, A. & Fei-Fei, L. Illuminating the dark spaces of healthcare with ambient intelligence. Nature 585, 193–202 (2020).
Zech, J. R. et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 15, e1002683 (2018).
Zou, J. & Schiebinger, L. AI can be sexist and racist — it’s time to make it fair. Nature 559, 324–326 (2018).
Perez, L. & Wang, J. The effectiveness of data augmentation in image classification using deep learning. Preprint at https://arxiv.org/abs/1712.04621v1 (2017).
Madani, A., Moradi, M., Karargyris, A. & Syeda-Mahmood, T. Semi-supervised learning with generative adversarial networks for chest X-ray classification with ability of data domain adaptation. In IEEE 15th International Symposium on Biomedical Imaging (ISBI) 1038–1042 (IEEE, 2018).
He, J. et al. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 25, 30–36 (2019).
Kelly, C. J., Karthikesalingam, A., Suleyman, M., Corrado, G. & King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 17, 195 (2019).
Rocher, L., Hendrickx, J. M. & de Montjoye, Y.-A. Estimating the success of re-identifications in incomplete datasets using generative models. Nat. Commun. 10, 3069 (2019).
Schwarz, C. G. et al. Identification of anonymous MRI research participants with face-recognition software. N. Engl. J. Med. 381, 1684–1686 (2019).
Chartsias, A., Joyce, T., Dharmakumar, R. & Tsaftaris, S. A. Adversarial image synthesis for unpaired multi-modal cardiac data. in Simulation and Synthesis in Medical Imaging (eds. Tsaftaris, S. A., Gooya, A., Frangi, A. F. & Prince, J. L.) 3–13 (Springer International Publishing, 2017).
Emami, H., Dong, M., Nejad-Davarani, S. P. & Glide-Hurst, C. K. Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med. Phys. https://doi.org/10.1002/mp.13047 (2018).
Jin, C.-B. et al. Deep CT to MR synthesis using paired and unpaired data. Sensors 19, 2361 (2019).
Bi, L., Kim, J., Kumar, A., Feng, D. & Fulham, M. In Molecular Imaging, Reconstruction and Analysis of Moving Body Organs, and Stroke Imaging and Treatment (eds. Cardoso, M. J. et al.) 43–51 (Springer International Publishing, 2017).
Ben-Cohen, A. et al. Cross-modality synthesis from CT to PET using FCN and GAN networks for improved automated lesion detection. Eng. Appl. Artif. Intell. 78, 186–194 (2019).
Armanious, K. et al. MedGAN: medical image translation using GANs. Comput. Med. Imaging Graph. 79, 101684 (2020).
Choi, H. & Lee, D. S. Alzheimer’s Disease Neuroimaging Initiative. Generation of structural MR images from amyloid PET: application to MR-less quantification. J. Nucl. Med. 59, 1111–1117 (2018).
Wei, W. et al. Learning myelin content in multiple sclerosis from multimodal MRI through adversarial training. In Medical Image Computing and Computer Assisted Intervention — MICCAI 2018 (eds. Frangi, A. F., Schnabel, J. A., Davatzikos, C., Alberola-López, C. & Fichtinger, G.) 514–522 (Springer Cham, 2018).
Pan, Y. et al. Synthesizing missing PET from MRI with cycle-consistent generative adversarial networks for Alzheimer’s disease diagnosis. In Medical Image Computing and Computer Assisted Intervention — MICCAI 2018 (eds. Frangi, A. F., Schnabel, J. A., Davatzikos, C., Alberola-López, C. & Fichtinger, G.) 455–463 (Springer Cham, 2018).
Welander, P., Karlsson, S. & Eklund, A. Generative adversarial networks for image-to-image translation on multi-contrast MR images - a comparison of CycleGAN and UNIT. Preprint at https://arxiv.org/abs/1806.07777v1 (2018).
Dar, S. U. H. et al. Image synthesis in multi-contrast MRI with conditional generative adversarial networks. IEEE Trans. Med. Imaging 38, 2375–2388 (2019).
Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In 2017 IEEE International Conference on Computer Vision (ICCV) (IEEE, 2017); https://doi.org/10.1109/iccv.2017.244
Maspero, M. et al. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys. Med. Biol. 63, 185001 (2018).
Olut, S., Sahin, Y.H., Demir, U., Unal, G. Generative adversarial training for MRA image synthesis using multi-contrast MRI. In PRedictive Intelligence in MEdicine. PRIME 2018. Lecture Notes in Computer Science (eds Rekik, I., Unal, G., Adeli, E. & Park, S.) (Springer Cham, 2018); https://doi.org/10.1007/978-3-030-00320-3_18
Chen, R. J., Lu, M. Y., Chen, T. Y., Williamson, D. F. K. & Mahmood, F. Synthetic data in machine learning for medicine and healthcare. Nat. Biomed. Eng. 5, 493–497 (2021).
Kanakasabapathy, M. K. et al. Adaptive adversarial neural networks for the analysis of lossy and domain-shifted datasets of medical images. Nat. Biomed. Eng. 5, 571–585 (2021).
Bowles, C., Gunn, R., Hammers, A. & Rueckert, D. Modelling the progression of Alzheimer’s disease in MRI using generative adversarial networks. In Medical Imaging 2018: Image Processing (eds. Angelini, E. D. & Landman, B. A.) 397– 407 (International Society for Optics and Photonics, 2018).
Ravi, D., Alexander, D.C., Oxtoby, N.P. & Alzheimer’s Disease Neuroimaging Initiative. Degenerative adversarial neuroImage nets: generating images that mimic disease progression. In Medical Image Computing and Computer Assisted Intervention — MICCAI 2019. Lecture Notes in Computer Science. (eds Shen, D. et al) 164–172 (Springer, 2019).
Borji, A. Pros and cons of GAN evaluation measures. Comput. Vis. Image Underst. 179, 41–65 (2019).
Vincent, J. Nvidia uses AI to make it snow on streets that are always sunny. The Verge https://www.theverge.com/2017/12/5/16737260/ai-image-translation-nvidia-data-self-driving-cars (2017).
Kairouz, P. et al. Advances and open problems in federated learning. Found. Trends Mach. Learn. https://doi.org/10.1561/2200000083 (2021)
McMahan, B., Moore, E., Ramage, D., Hampson, S. & Aguera y Arcas, B. Communication-efficient learning of deep networks from decentralized data. In Proceedings of the 20th International Conference on Artificial Intelligence and Statistics (eds. Singh, A. & Zhu, J.) 1273–1282 (ML Research Press, 2017).
Li, X. et al. Multi-site fMRI analysis using privacy-preserving federated learning and domain adaptation: ABIDE results. Med. Image Anal. 65, 101765 (2020).
Brisimi, T. S. et al. Federated learning of predictive models from federated Electronic Health Records. Int. J. Med. Inform. 112, 59–67 (2018).
Lee, J. et al. Privacy-preserving patient similarity learning in a federated environment: development and analysis. JMIR Med. Inform. 6, e20 (2018).
Dou, Q. et al. Federated deep learning for detecting COVID-19 lung abnormalities in CT: a privacy-preserving multinational validation study. npj Digit. Med. 4, 60 (2021).
Silva, S. et al. Federated learning in distributed medical databases: meta-analysis of large-scale subcortical brain data. In 2019 IEEE 16th International Symposium on Biomedical Imaging ISBI 2019 18822077 (IEEE, 2019).
Sheller, M. J., Reina, G. A., Edwards, B., Martin, J. & Bakas, S. Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. Brainlesion 11383, 92–104 (2019).
Sheller, M. J. et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 10, 12598 (2020).
Sarma, K. V. et al. Federated learning improves site performance in multicenter deep learning without data sharing. J. Am. Med. Inform. Assoc. 28, 1259–1264 (2021).
Li, W. et al. Privacy-preserving federated brain tumour segmentation. In Machine Learning in Medical Imaging (eds. Suk, H.-I., Liu, M., Yan, P. & Lian, C.) 133–141 (Springer International Publishing, 2019).
Shokri, R., Stronati, M., Song, C. & Shmatikov, V. Membership inference attacks against machine learning models. In IEEE Symposium on Security and Privacy SP 2017 3–18 (IEEE, 2017).
Fredrikson, M., Jha, S. & Ristenpart, T. Model inversion attacks that exploit confidence information and basic countermeasures. In Proceedings of the 22nd ACM SIGSAC Conference on Computer and Communications Security 1322–1333 (Association for Computing Machinery, 2015).
Zhang, C., Bengio, S., Hardt, M., Recht, B. & Vinyals, O. Understanding deep learning (still) requires rethinking generalization. Commun. ACM 64, 107–115 (2021).
Zhu, L., Liu, Z. & Han, S. Deep leakage from gradients. In Advances in Neural Information Processing Systems 32 (eds Wallach, H. et al.) Paper 8389 (Curran, 2019)
Abadi, M. et al. Deep learning with differential privacy. In Proceedings of the 2016 ACM SIGSAC Conference on Computer and Communications Security 308–318 (Association for Computing Machinery, 2016).
Brendan McMahan, H. et al. A general approach to adding differential privacy to iterative training procedures. Preprint at https://arxiv.org/abs/1812.06210v2 (2018).
McMahan, H. B., Ramage, D., Talwar, K. & Zhang, L. Learning differentially private recurrent language models. In ICLR 2018 Sixth International Conference on Learning Representations Paper 504 (ICLR, 2018).
Shokri, R. & Shmatikov, V. Privacy-preserving deep learning. In Proceedings of the 22nd ACM SIGSAC Conference on Computer and Communications Security 1310–1321 (Association for Computing Machinery, 2015).
Lyu, M., Su, D. & Li, N. Understanding the sparse vector technique for differential privacy. Proc. VLDB Endow. 10, 637–648 (2017).
Hitaj, B., Ateniese, G. & Perez-Cruz, F. Deep models under the GAN: information leakage from collaborative deep learning. In Proceedings of the 2017 ACM SIGSAC Conference on Computer and Communications Security 603–618 (Association for Computing Machinery, 2017).
Li, X., Huang, K., Yang, W., Wang, S. & Zhang, Z. On the convergence of FedAvg on Non-IID Data. In ICLR 2020 Eighth International Conference on Learning Representations Paper 261 (2020).
Smith, V., Chiang, C.-K., Sanjabi, M. & Talwalkar, A. S. Federated multi-task learning. In Advances in Neural Information Processing Systems 30 (eds Guyon, I. et al.) Paper 2307 (NeuIPS, 2017).
Xu, J. et al. Federated learning for healthcare informatics. J. Healthc. Inform. Res. 5, 1–19 (2021).
Huang, L. et al. LoAdaBoost: loss-based AdaBoost federated machine learning with reduced computational complexity on IID and non-IID intensive care data. PLoS ONE 15, e0230706 (2020).
Zhao, Y. et al. Federated learning with non-IID data. Preprint at https://arxiv.org/abs/1806.00582v1 (2018).
Torres-Soto, J. & Ashley, E. A. Multi-task deep learning for cardiac rhythm detection in wearable devices. npj Digit. Med. 3, 116 (2020).
Turakhia, M. P. et al. Rationale and design of a large-scale, app-based study to identify cardiac arrhythmias using a smartwatch: The Apple Heart Study. Am. Heart J. 207, 66–75 (2019).
Synced. Apple reveals design of its on-device ML system for federated evaluation and tuning SyncedReview https://syncedreview.com/2021/02/19/apple-reveals-design-of-its-on-device-ml-system-for-federated-evaluation-and-tuning (2021).
McMahan, B. & Ramage, D. Federated learning: collaborative machine learning without centralized training data Google AI Blog https://ai.googleblog.com/2017/04/federated-learning-collaborative.html (2017).
Chen, Y., Qin, X., Wang, J., Yu, C. & Gao, W. FedHealth: a federated transfer learning framework for wearable healthcare. IEEE Intell. Syst. 35, 83–93 (2020).
Ramage, D. & Mazzocchi, S. Federated analytics: collaborative data science without data collection Google AI Blog https://ai.googleblog.com/2020/05/federated-analytics-collaborative-data.html (2020).
Augenstein, S. et al. Generative models for effective ML on private, decentralized datasets. In ICLR 2020 Eighth International Conference on Learning Representations Paper 1448 (ICLR, 2020).
Pati, S. et al. The federated tumor segmentation (FeTS) challenge. Preprint at https://arxiv.org/abs/2105.05874v2 (2021).
Flores, M. Medical institutions collaborate to improve mammogram assessment AI with Nvidia Clara federated learning The AI Podcast https://blogs.nvidia.com/blog/2020/04/15/federated-learning-mammogram-assessment/ (2020).
Kannan, A., Chen, K., Jaunzeikare, D. & Rajkomar, A. Semi-supervised learning for information extraction from dialogue. In Proc. Interspeech 2018 2077–2081 (ISCA, 2018); https://doi.org/10.21437/interspeech.2018-1318
Chiu, C.-C. et al. Speech recognition for medical conversations. Preprint at https://arxiv.org/abs/1711.07274v2; https://doi.org/10.1093/jamia/ocx073 (2017).
Si, Y., Wang, J., Xu, H. & Roberts, K. Enhancing clinical concept extraction with contextual embeddings. J. Am. Med. Inform. Assoc. 26, 1297–1304 (2019).
Shin, H.-C. et al. Learning to read chest X-rays: recurrent neural cascade model for automated image annotation. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2016); https://doi.org/10.1109/cvpr.2016.274
Wang, X., Peng, Y., Lu, L., Lu, Z. & Summers, R. M. TieNet: text-image embedding network for common thorax disease classification and reporting in chest X-rays. In IEEE/CVF Conference on Computer Vision and Pattern Recognition 2018 (IEEE, 2018); https://doi.org/10.1109/cvpr.2018.00943
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1735–1780 (1997).
Cho, K. et al. Learning phrase representations using RNN encoder-decoder for statistical machine translation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) (eds Moschitti, A., Pang, B. & Daelemans, W.) 1724–1734 (Association for Computational Linguistics, 2014).
Lipton, Z. C., Kale, D. C., Elkan, C. & Wetzel, R. Learning to diagnose with LSTM recurrent neural networks. Preprint at https://arxiv.org/abs/1511.03677v7 (2015).
Choi, E., Bahadori, M. T., Schuetz, A., Stewart, W. F. & Sun, J. Doctor AI: predicting clinical events via recurrent neural networks. JMLR Workshop Conf. Proc. 56, 301–318 (2016).
Zhu, Paschalidis & Tahmasebi. Clinical concept extraction with contextual word embedding. Preprint at https://doi.org/10.48550/arXiv.1810.10566 (2018).
Cho, K., van Merriënboer, B., Bahdanau, D. & Bengio, Y. On the properties of neural machine translation: encoder–decoder approaches. In Proceedings of SSST-8, Eighth Workshop on Syntax, Semantics and Structure in Statistical Translation (eds Wu, D., Carpuat, M., Carreras, X. & Vecchi, E. M.) 103–111 (Association for Computational Linguistics, 2014).
Gehring, J., Auli, M., Grangier, D., Yarats, D. & Dauphin, Y. N. Convolutional sequence to sequence learning. In Proceedings of the 34th International Conference on Machine Learning (eds Precup, D. & Teh, Y. W.) 1243–1252 (PMLR, 2017).
Vaswani, A. et al. Attention is all you need. In Advances in Neural Information Processing Systems 30 (eds Guyon, I. et al.) Paper 3058 (Curran, 2017).
Bahdanau, D., Cho, K. H. & Bengio, Y. Neural machine translation by jointly learning to align and translate. In 3rd International Conference on Learning Representations ICLR 2015 (ICLR, 2015).
Luong, T., Pham, H. & Manning, C. D. Effective approaches to attention-based neural machine translation. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing (eds Màrquez, L., Callison-Burch, C. & Su, J.) 1412–1421 (Association for Computational Linguistics, 2015); https://doi.org/10.18653/v1/d15-1166
Sutskever, I., Vinyals, O. & Le, Q. V. Sequence to sequence learning with neural networks. In Advances in Neural Information Processing Systems 27 (eds Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N. & Weinberger, K. Q.) Paper 1610 (Curran, 2014).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. in Advances in Neural Information Processing Systems 25 (eds Bartlett, P. et al.) 1097–1105 (Curran, 2012).
Kiani, A. et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. npj Digit. Med. 3, 23 (2020).
Park, S.-M. et al. A mountable toilet system for personalized health monitoring via the analysis of excreta. Nat. Biomed. Eng. 4, 624–635 (2020).
Howard, J. & Ruder, S. Universal language model fine-tuning for text classification. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (eds Gurevych, I. & Miyao, Y.) 328–339 (Association for Computational Linguistics, 2018).
Peters, M. E. et al. Deep contextualized word representations. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (eds Walker, M., Ji, H. & Stent, A.) 2227–2237 (Association for Computational Linguistics, 2018).
Brown, T. et al. Language models are few-shot learners. In Advances in Neural Information Processing Systems 33 (eds. Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M. F. & Lin, H.) 1877–1901 (Curran, 2020).
Kenton, J. D. M.-W. C. & Toutanova, L. K. BERT: pre-training of deep bidirectional transformers for language understanding. in Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (eds Burstein, J., Doran, C. & Solorio, T.) 4171–4186 (Association for Computational Linguistics, 2019).
Mikolov, T., Chen, K., Corrado, G. & Dean, J. Efficient estimation of word representations in vector space. Preprint at https://arxiv.org/abs/1301.3781v3 (2013).
Pennington, J., Socher, R. & Manning, C. GloVe: global vectors for word representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (eds Moschitti, A., Pang, B., Daelemans, W.) 1532–1543 (Association for Computational Linguistics, 2014).
Alsentzer, E. et al. Publicly available clinical BERT embeddings. In Proceedings of the 2nd Clinical Natural Language Processing Workshop (eds Rumshisky, A., Roberts, K., Bethard, S. & Naumann, T.) 72–78 (Association for Computational Linguistics, 2019).
Huang, K., Altosaar, J. & Ranganath, R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. Preprint at https://arxiv.org/abs/1904.05342v3 (2019).
Peng, Y., Yan, S. & Lu, Z. Transfer learning in biomedical natural language processing: an evaluation of BERT and ELMo on ten benchmarking datasets. In Proceedings of the 18th BioNLP Workshop and Shared Task (eds Demner-Fushman, D., Bretonnel Cohen, K., Ananiadou, S. & Tsujii, J.) 58–65 (Association for Computational Linguistics, 2019).
Johnson, A. E. W. et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3, 160035 (2016).
Sharir, O., Peleg, B. & Shoham, Y. The cost of training NLP models: a concise overview. Preprint at https://arxiv.org/abs/2004.08900v1 (2020).
Lee, J. et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36, 1234–1240 (2020).
Beltagy, I., Lo, K. & Cohan, A. SciBERT: A pretrained language model for scientific text. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (eds Inui, K., Jiang, J., Ng, V. & Wan, X.) 3615–3620 (Association for Computational Linguistics, 2019).
Futoma, J., Morris, J. & Lucas, J. A comparison of models for predicting early hospital readmissions. J. Biomed. Inform. 56, 229–238 (2015).
Caruana, R. et al. Intelligible models for healthcare: predicting pneumonia risk and hospital 30-day readmission. In Proc. 21st ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 1721–1730 (Association for Computing Machinery, 2015).
Wagner, T. et al. Augmented curation of clinical notes from a massive EHR system reveals symptoms of impending COVID-19 diagnosis. Elife 9, e58227 (2020).
Eisman, A. S. et al. Extracting angina symptoms from clinical notes using pre-trained transformer architectures. AMIA Annu. Symp. Proc. 2020, 412–421 (American Medical Informatics Association, 2020).
Smit, A. et al. Combining automatic labelers and expert annotations for accurate radiology report labeling using BERT. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (eds Webber, B., Cohn, T., He, Y. & Liu, Y.) 1500–1519 (Association for Computational Linguistics, 2020).
Soni, S. & Roberts, K. Evaluation of dataset selection for pre-training and fine-tuning transformer language models for clinical question answering. In Proc. 12th Language Resources and Evaluation Conference 5532–5538 (European Language Resources Association, 2020).
Sezgin, E., Huang, Y., Ramtekkar, U. & Lin, S. Readiness for voice assistants to support healthcare delivery during a health crisis and pandemic. npj Digit. Med. 3, 122 (2020).
Sakthive, V., Kesaven, M. P. V., William, J. M. & Kumar, S. K. M. Integrated platform and response system for healthcare using Alexa. Int. J. Commun. Computer Technol. 7, 14–22 (2019).
Comstock, J. Buoy Health, CVS MinuteClinic partner to send patients from chatbot to care. mobihealthnews https://www.mobihealthnews.com/content/buoy-health-cvs-minuteclinic-partner-send-patients-chatbot-care (2018).
Razzaki, S. et al. A comparative study of artificial intelligence and human doctors for the purpose of triage and diagnosis. Preprint at https://doi.org/10.48550/arXiv.1806.10698 (2018).
Xiong, Y., Du, B. & Yan, P. Reinforced transformer for medical image captioning. In Machine Learning in Medical Imaging (eds. Suk, H.-I., Liu, M., Yan, P. & Lian, C.) 673–680 (Springer International Publishing, 2019).
Meng, Y., Speier, W., Ong, M. K. & Arnold, C. W. Bidirectional representation learning from transformers using multimodal electronic health record data to predict depression. IEEE J. Biomed. Health Inform. 25, 3121–3129 (2021).
Choi, E. et al. Learning the graphical structure of electronic health records with graph convolutional transformer. Proc. Conf. AAAI Artif. Intell. 34, 606–613 (2020).
Li, F. et al. Fine-tuning bidirectional encoder representations from transformers (BERT)–based models on large-scale electronic health record notes: an empirical study. JMIR Med. Inform. 7, e14830 (2019).
Rasmy, L., Xiang, Y., Xie, Z., Tao, C. & Zhi, D. Med-BERT: pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction. npj Digital Medicine 4, 86 (2021).
Shang, J., Ma, T., Xiao, C. & Sun, J. Pre-training of graph augmented transformers for medication recommendation. in Proceedings of the Twenty-Eighth International Joint Conference on Artificial Intelligence (ed. Kraus, S.) 5953–5959 (International Joint Conferences on Artificial Intelligence Organization, 2019); https://doi.org/10.24963/ijcai.2019/825
Li, Y. et al. BEHRT: transformer for electronic health records. Sci. Rep. 10, 7155 (2020).
Rao, S. et al. BEHRT-HF: an interpretable transformer-based, deep learning model for prediction of incident heart failure. Eur. Heart J. 41 (Suppl. 2), ehaa946.3553 (2020).
Qian, X. et al. Prospective assessment of breast cancer risk from multimodal multiview ultrasound images via clinically applicable deep learning. Nat. Biomed. Eng. 5, 522–532 (2021).
Xing, L., Giger, M. L. & Min, J. K. Artificial Intelligence in Medicine: Technical Basis and Clinical Applications (Academic Press, 2020).
Reisman, M. EHRs: the challenge of making electronic data usable and interoperable. P. T. 42, 572–575 (2017).
Cortés, R., Bonnaire, X., Marin, O. & Sens, P. Stream processing of healthcare sensor data: studying user traces to identify challenges from a big data perspective. Procedia Comput. Sci. 52, 1004–1009 (2015).
Zhang, F., Cao, J., Khan, S. U., Li, K. & Hwang, K. A task-level adaptive MapReduce framework for real-time streaming data in healthcare applications. Future Gener. Comput. Syst. 43–44, 149–160 (2015).
El Aboudi, N. & Benhlima, L. Big data management for healthcare systems: architecture, requirements, and implementation. Adv. Bioinformatics 2018, 4059018 (2018).
Ta, V.-D., Liu, C.-M. & Nkabinde, G. W. Big data stream computing in healthcare real-time analytics. In IEEE International Conference on Cloud Computing and Big Data Analysis (ICCCBDA) 37–42 (ieeexplore.ieee.org, 2016).
Data-Driven Healthcare Organizations Use Big Data Analytics for Big Gains White Paper (IBM Software, 2017); https://silo.tips/download/ibm-software-white-paper-data-driven-healthcare-organizations-use-big-data-analy
Futoma, J., Simons, M., Panch, T., Doshi-Velez, F. & Celi, L. A. The myth of generalisability in clinical research and machine learning in health care. Lancet Digit. Health 2, e489–e492 (2020).
Wang, X. et al. Inconsistent performance of deep learning models on mammogram classification. J. Am. Coll. Radiol. 17, 796–803 (2020).
Nestor, B., McDermott, M. B. A. & Boag, W. Feature robustness in non-stationary health records: caveats to deployable model performance in common clinical machine learning tasks. Preprint at https://doi.org/10.48550/arXiv.1908.00690 (2019).
Wu, E. et al. How medical AI devices are evaluated: limitations and recommendations from an analysis of FDA approvals. Nat. Med. https://doi.org/10.1038/s41591-021-01312-x (2021).
Barish, M., Bolourani, S., Lau, L. F., Shah, S. & Zanos, T. P. External validation demonstrates limited clinical utility of the interpretable mortality prediction model for patients with COVID-19. Nat. Mach. Intell. 3, 25–27 (2020).
Davis, S. E., Lasko, T. A., Chen, G., Siew, E. D. & Matheny, M. E. Calibration drift in regression and machine learning models for acute kidney injury. J. Am. Med. Inform. Assoc. 24, 1052–1061 (2017).
Wang, G. et al. A deep-learning pipeline for the diagnosis and discrimination of viral, non-viral and COVID-19 pneumonia from chest X-ray images. Nat. Biomed. Eng. 5, 509–521 (2021).
Ning, W. et al. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat. Biomed. Eng. 4, 1197–1207 (2020).
Koenecke, A. et al. Racial disparities in automated speech recognition. Proc. Natl Acad. Sci. USA 117, 7684–7689 (2020).
Abid, A., Farooqi, M. & Zou, J. Large language models associate muslims with violence. Nat. Mach. Intell. 3, 461–463 (2021).
Obermeyer, Z., Powers, B., Vogeli, C. & Mullainathan, S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 366, 447–453 (2019).
Adamson, A. S. & Smith, A. Machine learning and health care disparities in dermatology. JAMA Dermatol. 154, 1247–1248 (2018).
Han, S. S. et al. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J. Invest. Dermatol. 138, 1529–1538 (2018).
Subbaswamy, A., Adams, R. & Saria, S. Evaluating model robustness and stability to dataset shift. In Proceedings of The 24th International Conference on Artificial Intelligence and Statistics (eds. Banerjee, A. & Fukumizu, K.) 2611–2619 (PMLR, 2021).
Izzo, Z., Ying, L. & Zou, J. How to learn when data reacts to your model: performative gradient descent. In Proceedings of the 38th International Conference on Machine Learning (eds. Meila, M. & Zhang, T.) 4641–4650 (PMLR, 2021).
Ghorbani, A., Kim, M. & Zou, J. A Distributional framework for data valuation. In Proceedings of the 37th International Conference on Machine Learning (eds. Iii, H. D. & Singh, A.) 3535–3544 (PMLR, 2020).
Zhang, L., Deng, Z., Kawaguchi, K., Ghorbani, A. & Zou, J. How does mixup help with robustness and generalization? In International Conference on Learning Representations 2021 Paper 2273 (ICLR, 2021).
Schulam, P. & Saria, S. Can you trust this prediction? Auditing pointwise reliability after learning. In Proceedings of the Twenty-Second International Conference on Artificial Intelligence and Statistics (eds. Chaudhuri, K. & Sugiyama, M.) 1022–1031 (PMLR, 2019).
Liu, X. et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med. 26, 1364–1374 (2020).
Cruz Rivera, S. et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat. Med. 26, 1351–1363 (2020).
Nayak, P. Understanding searches better than ever before. Google The Keyword https://blog.google/products/search/search-language-understanding-bert/ (2019).
Baur, C., Albarqouni, S. & Navab, N. in OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis (eds Stoyanov, D. et al.) 260–267 (Springer International Publishing, 2018).
Kang, E., Koo, H. J., Yang, D. H., Seo, J. B. & Ye, J. C. Cycle-consistent adversarial denoising network for multiphase coronary CT angiography. Med. Phys. 46, 550–562 (2019).
Vig, J. A multiscale visualization of attention in the transformer model. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics: System Demonstrations (Costa-jussà, M. R. & Alfonseca, E.) 37–42 (Association for Computational Linguistics, 2019).
Acknowledgements
This work was supported in part by the National Institutes of Health via grants F30HL156478 (to A.Z.), R01CA227713 (to L.X.), R01CA256890 (to L.X.), P30AG059307 (to J.Z.), U01MH098953 (to J.Z.), P01HL141084 (to J.C.W), R01HL163680 (to J.C.W), R01HL130020 (to J.C.W), R01HL146690 (to J.C.W.) and R01HL126527 (to J.C.W.); by the National Science Foundation grant CAREER1942926 (to J.Z.); and by the American Heart Association grant 17MERIT3361009 (to J.C.W.). Figures were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
A.Z. and J.C.W. drafted the manuscript. All authors contributed to the conceptualization and editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.C.W. is a co-founder and scientific advisory board member of Greenstone Biosciences. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Pearse Keane, Faisal Mahmood and Hadi Shafiee for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, A., Xing, L., Zou, J. et al. Shifting machine learning for healthcare from development to deployment and from models to data. Nat. Biomed. Eng 6, 1330–1345 (2022). https://doi.org/10.1038/s41551-022-00898-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-022-00898-y
This article is cited by
-
Applying the UTAUT2 framework to patients’ attitudes toward healthcare task shifting with artificial intelligence
BMC Health Services Research (2024)
-
Generative models improve fairness of medical classifiers under distribution shifts
Nature Medicine (2024)
-
Algor-ethics: charting the ethical path for AI in critical care
Journal of Clinical Monitoring and Computing (2024)
-
Cervical cancer survival prediction by machine learning algorithms: a systematic review
BMC Cancer (2023)
-
Systematic review: fluid biomarkers and machine learning methods to improve the diagnosis from mild cognitive impairment to Alzheimer’s disease
Alzheimer's Research & Therapy (2023)