Abstract
Amyotrophic lateral sclerosis (ALS) is a heterogenous neurodegenerative disorder that affects motor neurons and voluntary muscle control1. ALS heterogeneity includes the age of manifestation, the rate of progression and the anatomical sites of symptom onset. Disease-causing mutations in specific genes have been identified and define different subtypes of ALS1. Although several ALS-associated genes have been shown to affect immune functions2, whether specific immune features account for ALS heterogeneity is poorly understood. Amyotrophic lateral sclerosis-4 (ALS4) is characterized by juvenile onset and slow progression3. Patients with ALS4 show motor difficulties by the time that they are in their thirties, and most of them require devices to assist with walking by their fifties. ALS4 is caused by mutations in the senataxin gene (SETX). Here, using Setx knock-in mice that carry the ALS4-causative L389S mutation, we describe an immunological signature that consists of clonally expanded, terminally differentiated effector memory (TEMRA) CD8 T cells in the central nervous system and the blood of knock-in mice. Increased frequencies of antigen-specific CD8 T cells in knock-in mice mirror the progression of motor neuron disease and correlate with anti-glioma immunity. Furthermore, bone marrow transplantation experiments indicate that the immune system has a key role in ALS4 neurodegeneration. In patients with ALS4, clonally expanded TEMRA CD8 T cells circulate in the peripheral blood. Our results provide evidence of an antigen-specific CD8 T cell response in ALS4, which could be used to unravel disease mechanisms and as a potential biomarker of disease state.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
CITE-seq, SMART-seq and RNA-seq datasets have been deposited online in the Gene Expression Omnibus (GEO) under the accession codes GSE180410, GSE199663, GSE199664 and GSE199666. Source data are provided with this paper.
Change history
09 August 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41586-022-05184-0
References
Brown, R. H. & Al-Chalabi, A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172 (2017).
McCauley, M. E. & Baloh, R. H. Inflammation in ALS/FTD pathogenesis. Acta Neuropathol. 137, 715–730 (2019).
Grunseich, C. et al. Clinical and molecular aspects of senataxin mutations in amyotrophic lateral sclerosis 4. Ann. Neurol. 87, 547–555 (2020).
Boillee, S. et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312, 1389–1392 (2006).
Yamanaka, K. et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 11, 251–253 (2008).
Butovsky, O. et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J. Clin. Invest. 122, 3063–3087 (2012).
McCauley, M. E. et al. C9orf72 in myeloid cells suppresses STING-induced inflammation. Nature 585, 96–101 (2020).
O’Rourke, J. G. et al. C9orf72 is required for proper macrophage and microglial function in mice. Science 351, 1324–1329 (2016).
Atanasio, A. et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci. Rep. 6, 23204 (2016).
Burberry, A. et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci. Transl. Med. 8, 347ra393 (2016).
Oakes, J. A., Davies, M. C. & Collins, M. O. TBK1: a new player in ALS linking autophagy and neuroinflammation. Mol. Brain 10, 5 (2017).
Xiao, Y. et al. The kinase TBK1 functions in dendritic cells to regulate T cell homeostasis, autoimmunity, and antitumor immunity. J. Exp. Med. 214, 1493–1507 (2017).
Becherel, O. J. et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 9, e1003435 (2013).
Miller, M. S. et al. Senataxin suppresses the antiviral transcriptional response and controls viral biogenesis. Nat. Immunol. 16, 485–494 (2015).
Bennett, C. L. et al. Senataxin mutations elicit motor neuron degeneration phenotypes and yield TDP-43 mislocalization in ALS4 mice and human patients. Acta Neuropathol. 136, 425–443 (2018).
Rathinam, V. A. et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by Gram-negative bacteria. Cell 150, 606–619 (2012).
Yu, C. H. et al. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell 183, 636–649 (2020).
Gate, D. et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404 (2020).
Gate, D. et al. CD4+ T cells contribute to neurodegeneration in Lewy body dementia. Science 374, 868–874 (2021).
Lindestam Arlehamn, C. S. et al. α-Synuclein-specific T cell reactivity is associated with preclinical and early Parkinson's disease. Nat. Commun. 11, 1875 (2020).
Sulzer, D. et al. T cells from patients with Parkinson’s disease recognize α-synuclein peptides. Nature 546, 656–661 (2017).
Hashimoto, M. et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu. Rev. Med. 69, 301–318 (2018).
Baruch, K. et al. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer’s disease. Nat. Med. 22, 135–137 (2016).
Schenkel, J. M., Fraser, K. A. & Masopust, D. Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. J. Immunol. 192, 2961–2964 (2014).
Baron, J. L., Madri, J. A., Ruddle, N. H., Hashim, G. & Janeway, C. A. Jr Surface expression of alpha 4 integrin by CD4 T cells is required for their entry into brain parenchyma. J. Exp. Med. 177, 57–68 (1993).
Young, K. G., Maclean, S., Dudani, R., Krishnan, L. & Sad, S. CD8+ T cells primed in the periphery provide time-bound immune-surveillance to the central nervous system. J. Immunol. 187, 1192–1200 (2011).
Campisi, L. et al. Apoptosis in response to microbial infection induces autoreactive TH17 cells. Nat. Immunol. 17, 1084–1092 (2016).
Ma, A., Koka, R. & Burkett, P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu. Rev. Immunol. 24, 657–679 (2006).
Sekine, T. et al. TOX is expressed by exhausted and polyfunctional human effector memory CD8+ T cells. Sci. Immunol. 5, eaba7918 (2020).
Markovic-Plese, S., Cortese, I., Wandinger, K. P., McFarland, H. F. & Martin, R. CD4+CD28− costimulation-independent T cells in multiple sclerosis. J. Clin. Invest. 108, 1185–1194 (2001).
Schmidt, D., Goronzy, J. J. & Weyand, C. M. CD4+ CD7- CD28- T cells are expanded in rheumatoid arthritis and are characterized by autoreactivity. J. Clin. Invest. 97, 2027–2037 (1996).
Gearty, S. V. et al. An autoimmune stem-like CD8 T cell population drives type 1 diabetes. Nature 602, 156–161 (2022).
Page, N. et al. Persistence of self-reactive CD8+ T cells in the CNS requires TOX-dependent chromatin remodeling. Nat. Commun. 12, 1009 (2021).
Dulken, B. W. et al. Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210 (2019).
Derecki, N. C. et al. Regulation of learning and memory by meningeal immunity: a key role for IL-4. J. Exp. Med. 207, 1067–1080 (2010).
Filiano, A. J., Gadani, S. P. & Kipnis, J. How and why do T cells and their derived cytokines affect the injured and healthy brain? Nat. Rev. Neurosci. 18, 375–384 (2017).
Kipnis, J. Multifaceted interactions between adaptive immunity and the central nervous system. Science 353, 766–771 (2016).
Chiffelle, J. et al. T-cell repertoire analysis and metrics of diversity and clonality. Curr. Opin. Biotechnol. 65, 284–295 (2020).
Oldrini, B. et al. Somatic genome editing with the RCAS-TVA-CRISPR-Cas9 system for precision tumor modeling. Nat. Commun. 9, 1466 (2018).
Kahles, A. et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224 (2018).
Ren, P. et al. Alternative splicing: a new cause and potential therapeutic target in autoimmune disease. Front. Immunol. 12, 713540 (2021).
Kurepa, Z., Su, J. & Forman, J. Memory phenotype of CD8+ T cells in MHC class Ia-deficient mice. J. Immunol. 170, 5414–5420 (2003).
Korobeynikov, V. A., Lyashchenko, A. K., Blanco-Redondo, B., Jafar-Nejad, P. & Shneider, N. A. Antisense oligonucleotide silencing of FUS expression as a therapeutic approach in amyotrophic lateral sclerosis. Nat. Med. 28, 104–116 (2022).
Gurney, M. E. et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775 (1994).
Tu, P. H. et al. Transgenic mice carrying a human mutant superoxide dismutase transgene develop neuronal cytoskeletal pathology resembling human amyotrophic lateral sclerosis lesions. Proc. Natl Acad. Sci. USA 93, 3155–3160 (1996).
Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999).
Patil, V. S. et al. Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol. 3, eaan8664 (2018).
Emerson, R. O. et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat. Genet. 49, 659–665 (2017).
Tickotsky, N., Sagiv, T., Prilusky, J., Shifrut, E. & Friedman, N. McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics 33, 2924–2929 (2017).
Weekes, M. P., Wills, M. R., Mynard, K., Carmichael, A. J. & Sissons, J. G. The memory cytotoxic T-lymphocyte (CTL) response to human cytomegalovirus infection contains individual peptide-specific CTL clones that have undergone extensive expansion in vivo. J. Virol. 73, 2099–2108 (1999).
Latorre, D. et al. T cells in patients with narcolepsy target self-antigens of hypocretin neurons. Nature 562, 63–68 (2018).
Campisi, L., Cummings, R. J. & Blander, J. M. Death-defining immune responses after apoptosis. Am. J. Transplant. 14, 1488–1498 (2014).
Weyand, C. M. & Goronzy, J. J. The immunology of rheumatoid arthritis. Nat. Immunol. 22, 10–18 (2021).
Garretti, F., Agalliu, D., Lindestam Arlehamn, C. S., Sette, A. & Sulzer, D. Autoimmunity in Parkinson’s disease: the role of α-synuclein-specific T cells. Front. Immunol. 10, 303 (2019).
Rabin, B. A. et al. Autosomal dominant juvenile amyotrophic lateral sclerosis. Brain 122, 1539–1550 (1999).
Lamoreaux, L., Roederer, M. & Koup, R. Intracellular cytokine optimization and standard operating procedure. Nat. Protoc. 1, 1507–1516 (2006).
Campisi, L. In vitro antigen-presentation assay for self- and microbial-derived antigens. Bio. Protoc. 7, e2307 (2017).
Korin, B., Dubovik, T. & Rolls, A. Mass cytometry analysis of immune cells in the brain. Nat. Protoc. 13, 377–391 (2018).
Tan, Y. S. & Lei, Y. L. Isolation of tumor-infiltrating lymphocytes by ficoll-paque density gradient centrifugation. Methods Mol. Biol. 1960, 93–99 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Shen, S. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl Acad. Sci. USA 111, E5593–E5601 (2014).
Low, D. SPLINTER: splice interpreter of transcripts. R version 1.4.0 https://github.com/dianalow/SPLINTER/ (2017).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Borcherding, N., Bormann, N. L. & Kraus, G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res. 9, 47 (2020).
Cuadrado-Castano, S. et al. Enhancement of the proapoptotic properties of newcastle disease virus promotes tumor remission in syngeneic murine cancer models. Mol. Cancer Ther. 14, 1247–1258 (2015).
Martinez, N. M. & Lynch, K. W. Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol. Rev. 253, 216–236 (2013).
Acknowledgements
We thank N. Zhao and S. Zheng from the I.M. laboratory for critical inputs of the study; all the teams at ISMMS for sharing reagents; E. Drapeau for teaching the use of the rotarod; T. Kraus for use of the Luminex 100/200 plate reader; S. Cuadrado-Castano and A. Javaher for help with the B16 melanoma model; the Flow Cytometry and Mouse facilities for training and animal care, and the NGS Platform of the Center for Advanced Genomics Technology for SMART-sequencing; T. Dawson, D. D’souza, B. Upadhyaya and all the team of the Human Immune Monitoring Center for CITE-sequencing and mass cytometry acquisition; and A. Soto, M. Garcia-Barros and the Biorepository and Pathology Dean’s CoRE for assistance with histology. We are grateful to L. Manganaro, J. Moretti and A. Chan for their help in the preliminary phase of the project; M. J. Miller and M. F. Lavin for Setx-deficient mice; the Genomic and Microarray Core Facility of University of Texas Southwestern Medical Center for the autoantigen microarray analysis ; the Duke Electron Microscopy Service of the Duke University Pathology Core for ultrathin sections of axons; The Johns Hopkins ALS Postmortem Core and K. Wilsbach and K. Gallo for providing post-mortem human tissues; and the New York Blood Center for providing buffy coats. We are indebted to ALS4.org for help and support. This work was supported by The Burroughs Wellcome Fund 1017892 (I.M.); the Chan Zuckerberg Initiative Neurodegeneration Challenge Network: the Ben Barres Early Career Acceleration Awards (to I.M., 2018-191895, and N.J., 2018-191856 (5022)) and the Collaborative Science Award (to A.R.L.S., 108313-00002); the Hirschl Young Investigator fellowship (I.M.); National Institutes of Health grants U01AI150748, R01AI143840 and R01AI168130 (I.M.), T32 AI007647 and T32 HD075735 (Y.F.), R33CA225539 and R01AG067581 (N.J.), R35 NS122140 (A.R.L.S.), T32 AG00096 (F.J.A.), 5R01NS106236 (N.A.S.), R01HL153974 and U01AI150748 (M.B.); the start-up fund of the School of Engineering and Applied Science at University of Pennsylvania (N.J.); and ARISLA Italy, ALS Association ALSA, USA, and the Italian Ministry of Health CCM2011 (C.L.). M.S. received support from the Seve Ballesteros Foundation. J.S.Y.H. is supported by the Charles H Revson Foundation Biomedical Science Fellowship. M.G.-F. is a recipient of a post-doctoral fellowship from the Fundación Alfonso Martín Escudero.
Author information
Authors and Affiliations
Contributions
L.C. and I.M. conceived, designed and directed the study. S.D.R., M.B., M.M.S., M.S., L.W.O., C.L., N.A.S., N.J. and A.R.L.S. provided conceptual and experimental guidance. L.C. conducted all experiments with the assistance of A.G. and F.J.A. (histology); V.A.K. (ALS6 mouse model); L.M. (blood collection and isolation of PBMCs); J.S.Y.H., Y.F. and C.B. (mouse dissection); J.P. (mouse TCR sequencing), J.S.Y.H., M.G.-F. and M.J.-A. (tumour models); and C.L.B. (preliminary analyses on SetxL389S+/− mice). M.J.-A. and M.S. generated and provided tumour-derived NSCs (glioma model). L.C. analysed and visualized the data. S.C. performed and visualized CITE-seq and SMART-seq analyses. J.S.Y.H. and D.T. performed and visualized splicing analyses. X.M. conceived and performed the axon counting by Gaussian filters, under the supervision of Z.A.F. L.C. administered the project. I.M. acquired funding. L.C. wrote the paper with edits from I.M., A.R.L.S. and S.C., and with feedback from all of the authors.
Corresponding authors
Ethics declarations
Competing interests
C.L. is the scientific director of the company NEMO Lab, and he served on a scientific advisory board for Mitsubishi Tanabe Pharma Europe, Cytokinetics, Neuraltus and Italfarmaco. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Aaron Gitler and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Characterization of KI mice.
a, Average latency to fall (sec) from an accelerating rotarod of WT and KI mice tested at 6 and 8 months of age (2 independent experiments, n = 11 (WT 6 months and KI 8 months) and n = 13 (WT 8 months and KI 6 months) individual mice per group; mean +/− s.e.m.). P value calculated using the one-way Repeated Measures ANOVA (**P = 0.0012) and Bonferroni’s post-test. (b,c) Bone marrow derived dendritic cells (BMDCs) from Setx WT and KI (b,c), and HET and knockout (KO) (b) mice were infected with Citrobacter rodentium (multiplicity of infection = 10, b) or stimulated with 5 μg/ml of cGAMP (c). Bar graphs show the relative induction of the indicated transcripts by RT–qPCR analysis at 2 h post-infection or cGAMP stimulation in KO and KI cells normalized to their controls (2 independent experiments, n = 4 mice/group, mean +/− s.e.m.). No expression of the indicated genes was detected in uninfected cells. d, Concentration of the indicated cytokines measured by Luminex Assay in the serum of KI and WT (8-9 months old) mice (1 out of 2 independent experiments, n = 6 (WT) and n = 5 (KI) mice/group, mean +/− s.e.m.). e, RT–qPCR analysis for the indicated inflammatory genes in the spinal cord of 10 months old WT and KI mice (Log2 relative expression against Actb, 2 independent experiments, n = 5 mice/group, mean +/− s.e.m.). f, Absolute numbers of indicated immune cell subsets in the spleen of 8-9 month-old KI and WT mice. pDC: plasmacytoid dendritic cells; cDC: conventional dendritic cells. g, Expression of the activation marker CD86 in cDC and CD11b+ cells from the spleen of 8-9 months old WT and KI mice. (f,g) 2 independent experiments, n = 4 mice/group, mean +/− s.e.m.
Extended Data Fig. 2 Autoantibody secretion in KI and wild-type mice.
Heat maps showing an unsupervised hierarchical clustering based on IgM (top) and IgG (bottom) reactivity against 128 self-antigens in the serum of 10–12-month-old KI and WT mice (n = 5 mice/group).
Extended Data Fig. 3 Immune cell subsets in WT and KI mice; related to Figure 1.
a,b, Peripheral blood cells from 8-month-old WT and KI mice were analysed by mass cytometry. a, t-SNE plots after gating on live CD45high (top) and CD19−NK1.1−CD11b−CD11c−TCRβ+ (bottom). b, Frequency of PD-1+ expressing cells after gating on CD44+ CD8+ T cells. Three mice/group were pooled for mass cytometry analysis. c, Gating strategy for flow cytometry analysis of CD8 T cells in the CNS. d, Frequency of PD-1+ CD8 T cells in the spleen and lungs of WT and KI mice at the indicated ages (2 independent experiments, 4 mice/group; mean +/− s.e.m.). e, Frequency of PD-1+ (left) and Foxp3+ (right) CD4 T cells in the peripheral blood of WT and KI mice at the indicated ages (3 independent experiments, n = 6 (left) and 2 independent experiments, n = 4 (8 months) and n = 6 (right) individual mice/group. SB=super bright.
Extended Data Fig. 4 PD-1+ CD8 T cells are effector memory T cells activated in an antigen manner.
a, Expression of indicated surface markers in CD8 T cells from the spinal cord (top), brain (middle) and peripheral blood (bottom) of 10–12-month-old KI mice after gating on PD-1+ and PD-1− CD8 T cells. Histograms are representative of 3 independent experiment (n = 6 mice). b, Median fluorescence intensity (MFI) of the indicated markers in CD44+CD62L− PD-1+ and PD-1− CD8 T cells isolated from the spinal cord (top) and blood (bottom) of 10–12-month-old KI mice (2 independent experiments, n = 6 (blood TIGIT and TOX), n = 5 (spinal cord TIGIT and TOX), n = 4 (TCF-1) individual mice/group, mean +/− s.e.m.). P value calculated using a two-tailed unpaired t-test with Welch’s correction (spinal cord: ***P = 0.0007; *P = 0.045; **P = 0.0066; blood: ***P = 0.0004 (TOX); ***P = 0.0002 (TIGIT and TCF-1). c, KI mice were crossed to OT-I mice and the offspring analysed at 10 months of age. Dot plots from spinal cords, SC (left), and bar graphs (right) show the frequency of PD-1+ among OVA specific (gated on Vα2+Vβ5+) and non-TCR transgenic (gated on Vα2−) CD8 T cells (3 independent experiments, n = 6, brain and spinal cord, and n = 5, blood, individual mice; mean+/− s.e.m.). P value calculated using a two-tailed unpaired t test with Welch’s correction. **P = 0.0031 (brain); ***P = 0.0004 (SC); ***P = 0.0003 (blood). eF=eFluor.
Extended Data Fig. 5 Anti-tumour immunity and alternative splicing in KI mice.
a, Kinetics of tumour growth in 10–12-month-old KI and WT mice subcutaneously injected with B16-F10 cells. Two independent experiments, n = 10 individual mice/group; mean+/− s.e.m. ns = not significant, P value calculated using a two-tailed Wilcoxon matched-pairs signed rank test. b, Scans of CD8 staining inside glioma isolated from 3 individual WT and KI from 2 independent experiments at day 27 post-tumour detection. c, Spinal cords from 10 to 12-month-old WT (n = 4) and KI (n = 3) mice (top) and in vitro activated WT CD8 T cells (n = 3) (bottom) were analysed by RNA-seq. WT CD8 T cells were used as positive control because increased AS after T cell activation has been reported73. Bar graphs show transcript counts of significantly changed (| deltaPSI | > 0.15, p < 0.05, FDR < 0.05) AS events using mm10 annotation. n = individual mice per group. SE, exon skipping; RI, intron retention; MXE, mutually exclusive exons; A5SS, alternative 5′ splice site; A3SS, alternative 3′ splice site.
Extended Data Fig. 6 Immune cell subsets in WT and KI mice; related to Figure 3.
a, Schematic of bone marrow cell transplantation experiments. b, Representative dot plots of the frequency of CD45.2 (recipient) and CD45.1 (donor) cells in the peripheral blood of chimeric mice 8 weeks after irradiation and reconstitution. c, Average of motor neuron area calculated in the lumbar region of spinal cord sections. n = 5 mice/group, mean +/− s.e.m. P value calculated using one-way ANOVA Kruskal-Wallis test and Dunn’s post-hoc test (*P = 0.0002). d, Axon areas (left) and number of large calibre axons (right) in the sciatic nerve. n = 6 (WT–WT, KI–KI, KI–WT) and 7 (WT–KI) mice per group; mean +/− s.e.m. P value calculated using one-way ANOVA Kruskal-Wallis test and Dunn’s post-hoc test (*P = 0.026, ns=not statistically significant). Chimeric mice in (c,d) were analysed at 12-13 months after reconstitution. e, Peripheral blood cells from two mice of the indicated group were pooled and analysed by mass cytometry. Left, Details of markers and cell subsets identified by SPADE (spanning-tree progression analysis for density-normalized events) analysis (WT–WT group is shown). Right, SPADE analysis showing the distribution of immune cells into 4 main populations. Size and colour of circles are dependent on the number of cells and the median intensity of expression for each marker, respectively. Red circles highlight PD-1+ CD8 T cells.
Extended Data Fig. 7 Autoantibody secretion in KI–KI and WT–WT chimeric mice.
Heat maps showing an unsupervised hierarchical clustering based on IgM (top) and IgG (bottom) reactivity against 128 self-antigens in the serum of chimeric mice at 12 months after reconstitution (n = 3 WT–WT and n = 2 KI–KI individual chimeras).
Extended Data Fig. 8 Absence of T cell signature in FusP517L/WT and SOD1G93A mice.
a,d, Cell suspensions from brain, spinal cord, spleen, and peripheral blood of 22–24 month-old FusP517L/WT and FusWT/WT littermates were analysed by spectral flow cytometry. a–c, t-SNE of total live CD45+ cells (a) and frequency of TCRβ+ CD8+ and CD4+ T cells expressing CD62L, CD44, PD-1+ and Foxp3 (b,c) from the spinal cord of FusP517L/WT and FusWT/WT mice after concatenation of all of the 5 samples/group. d, Frequency of CD8 T cell subsets in the indicated organs from the 2 genotypes (2 independent experiments, 5 individual mice/group; mean +/− s.e.m.). e, Frequency of CD8 T cell subsets in the indicated organs from 3-4 month-old non-carrier and SOD1G93A mice by spectral flow cytometry analysis (2 independent experiments, n = 5 (spleen), n = 6 (brain, WT blood) and n = 7 individual mice/group; mean +/− s.e.m.). N= naïve, CD62L+CD44−; CM= central memory, CD62L+CD44+; EM= effector memory CD62L−CD44+. T cells are gated on live CD45high CD19−NK1.1−CD11b−CD11c−TCRγδ−TCRβ+ CD8+ or CD4+ cells.
Extended Data Fig. 9 Immune cell subsets in WT and KI mice; related to Fig. 4.
a, PBMCs from age-matched control individuals and patients were analysed by spectral flow cytometry. Frequencies of naïve (CD45RA+ CD45RO−CD27+CD28+) and central memory (TCM, CD45RO+ CD45RA− CD28+) were determined after gating on CCR7+ CD8 (left) or CD4 (right) T cells. Frequencies of effector (TEff, CD45RA+/− CD45RO+/− CD27+CD28+), effector memory (TEM, CD45RA− CD45RO+ CD27+CD28+) and terminally differentiated effector memory (TEMRA, CD45RO− CD45RA+ CD28−) were determined after gating on CCR7− CD8 or CD4 T cells. T cells were gated on live CD19−CD16− CD56−CD3+ cells. Each dot represents one individual donor and graph show results from 2 independent experiments (n = 5 ALS4 and n = 10 controls). mean +/− s.e.m. P value calculated for each T cell subset using Unpaired two-tailed t test with Welch’s correction (Teff: ***P = 0.0002; TEMRA: ***P = 0.0067). ns= not significant. b, Levels of expression of TOX, TIGIT and TCF-1 in the indicated CD8 T cell subsets from the peripheral blood of patients with ALS4 (n = 4) and age-matched controls (n = 8). Two independent experiments, mean +/− s.e.m. P value calculated by comparing TEMRA CD8 T cells from patients and control individuals pool together to the other subsets using one-way ANOVA and Tukey post-hoc test. c,d, UMAP projections of 20,000 single cells from control individuals (n = 3) and patients with ALS4 (n = 3) coloured by cluster (c) or by cell subsets (d) are shown on the left and Z-score normalized mean expression of differentially expressed genes in the indicated clusters (c) or CD8 T cell subsets (d) are shown on the right.
Extended Data Fig. 10 Phenotype of CD8 T cells from patients with ALS4 and control individuals.
a,b, UMAP projections of identified clusters showing differential expression of surface markers (by oligo-conjugated antibodies) (a) and of the top genes (b) in ALS4 versus control donors. c, Spectral flow cytometry analysis of the 6 samples used for CITE-seq. Histograms show the frequency of CCR7+ CD8 T cells and dot plots the proportion of the indicated subsets after gating on CCR7pos or neg CD8 T cells as indicated.
Extended Data Fig. 11 Clonal expansion and antigen specificity of TEMRA CD8 T cells from patients with ALS4.
a–c, CD8 T cells from controls and patients with ALS4 were analysed by CITE-seq. a, Donut plots depicting T cell clonality per CD8 T cell subset and per group. b, Donut plots indicating the number of TEMRA CD8 T cells for each indicated clonal category. Each patient and the corresponding age-matched controls are individually shown. c, Z-score normalized mean expression of differentially expressed surface markers (by oligo-conjugated antibodies) in “High” (clonality > 20) versus “Low” expanded TEMRA CD8 T cells from patients with ALS4. “High” and “Low” clones have been subsampled for equal representation on the heat map. d, PBMCs from 2 patients with ALS4 and 4 age-matched control individuals were analysed by spectral flow cytometry using a mixture of fluorophore-conjugated antibodies against 24 human TCR-Vβ. Graph bars show the frequencies of the top 5 TCR VB expressed by TEMRA CD8 T cells in each donor. Top 5 VB chains are indicated in the table below the graph. e, Concentration of IFNγ in the supernatant of PBMCs from control individuals (n = 10) and patients with ALS4 (n = 4) stimulated for 6 days with 15mers overlapping TDP-43 (106 peptides), Senataxin (703 peptides divided in 2 sub-pools), or with a pool of 80 pathogen-derived peptides. Two independent experiments, mean +/− s.e.m.
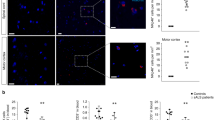
Extended Data Fig. 12 CD8 T cells are detected in the ventral horn of lumbar spinal cords from patients with ALS4.
CD8 T cell staining in the indicated post-mortem paraffin-embedded tissues from patients with ALS4 and controls. For spinal cord sections, scans show the ventral horn. Narrows indicate CD8 T cells. n = 2.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 5 & 9, and legends for Supplementary Tables 1–8.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Campisi, L., Chizari, S., Ho, J.S.Y. et al. Clonally expanded CD8 T cells characterize amyotrophic lateral sclerosis-4. Nature 606, 945–952 (2022). https://doi.org/10.1038/s41586-022-04844-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04844-5
This article is cited by
-
Comparison of T-cell receptor diversity of people with myalgic encephalomyelitis versus controls
BMC Research Notes (2024)
-
Senataxin helicase, the causal gene defect in ALS4, is a significant modifier of C9orf72 ALS G4C2 and arginine-containing dipeptide repeat toxicity
Acta Neuropathologica Communications (2023)
-
Opinion: more mouse models and more translation needed for ALS
Molecular Neurodegeneration (2023)
-
Senataxin and R-loops homeostasis: multifaced implications in carcinogenesis
Cell Death Discovery (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.