Abstract
Extracellular acidification occurs in inflamed tissue and the tumor microenvironment; however, a systematic study on how pH sensing contributes to tissue homeostasis is lacking. In the present study, we examine cell type-specific roles of the pH sensor G protein-coupled receptor 65 (GPR65) and its inflammatory disease-associated Ile231Leu-coding variant in inflammation control. GPR65 Ile231Leu knock-in mice are highly susceptible to both bacterial infection-induced and T cell-driven colitis. Mechanistically, GPR65 Ile231Leu elicits a cytokine imbalance through impaired helper type 17 T cell (TH17 cell) and TH22 cell differentiation and interleukin (IL)-22 production in association with altered cellular metabolism controlled through the cAMP–CREB–DGAT1 axis. In dendritic cells, GPR65 Ile231Leu elevates IL-12 and IL-23 release at acidic pH and alters endo-lysosomal fusion and degradation capacity, resulting in enhanced antigen presentation. The present study highlights GPR65 Ile231Leu as a multistep risk factor in intestinal inflammation and illuminates a mechanism by which pH sensing controls inflammatory circuits and tissue homeostasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The scRNA-seq data generated during the present study are available under Gene Expression Omnibus accession no. GSE182767. The reference mm10 mouse transcriptome was obtained from GENCODE: GCF_000001635.26. Source data are provided with this paper.
Code availability
Code used for the present study is available at https://gitlab.com/xavier-lab-computation/public/gpr65-scrnaseq.
References
Wu, H. et al. T-cells produce acidic niches in lymph nodes to suppress their own effector functions. Nat. Commun. 11, 4113 (2020).
Ihara, Y. et al. The G protein-coupled receptor T-cell death-associated gene 8 (TDAG8) facilitates tumor development by serving as an extracellular pH sensor. Proc. Natl Acad. Sci. USA 107, 17309–17314 (2010).
Levin, L. R. & Buck, J. Physiological roles of acid-base sensors. Annu. Rev. Physiol. 77, 347–362 (2015).
Ananthakrishnan, A. N. et al. Environmental triggers in IBD: a review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol. 15, 39–49 (2018).
de Souza, H. S. & Fiocchi, C. Immunopathogenesis of IBD: current state of the art. Nat. Rev. Gastroenterol. Hepatol. 13, 13–27 (2016).
Kamada, N., Seo, S. U., Chen, G. Y. & Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321–335 (2013).
Hardin, M. et al. The clinical and genetic features of COPD-asthma overlap syndrome. Eur. Respir. J. 44, 341–350 (2014).
Wirasinha, R. C. et al. GPR65 inhibits experimental autoimmune encephalomyelitis through CD4+ T cell independent mechanisms that include effects on iNKT cells. Immunol. Cell Biol. 96, 128–136 (2018).
International Multiple Sclerosis Genetics Consortium et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011).
Lassen, K. G. et al. Genetic coding variant in GPR65 alters lysosomal pH and links lysosomal dysfunction with colitis risk. Immunity 44, 1392–1405 (2016).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Tcymbarevich, I. et al. Lack of the pH-sensing receptor TDAG8 [GPR65] in macrophages plays a detrimental role in murine models of inflammatory bowel disease. J. Crohns Colitis 13, 245–258 (2019).
Xie, L. et al. pH and proton sensor GPR65 determine susceptibility to atopic dermatitis. J. Immunol. 207, 101–109 (2021).
Wang, Y. et al. The proton-activated receptor GPR4 modulates intestinal inflammation. J. Crohns Colitis 12, 355–368 (2018).
de Valliere, C. et al. G Protein-coupled pH-sensing receptor OGR1 Is a regulator of intestinal inflammation. Inflamm. Bowel Dis. 21, 1269–1281 (2015).
Basu, R. et al. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity 37, 1061–1075 (2012).
Ishigame, H. et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30, 108–119 (2009).
Harbour, S. N., Maynard, C. L., Zindl, C. L., Schoeb, T. R. & Weaver, C. T. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc. Natl Acad. Sci. USA 112, 7061–7066 (2015).
Ivanov, I. I. et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Plank, M. W. et al. Th22 cells form a distinct Th lineage from Th17 cells in vitro with unique transcriptional properties and Tbet-dependent Th1 plasticity. J. Immunol. 198, 2182–2190 (2017).
Wang, C. et al. CD5L/AIM regulates lipid biosynthesis and restrains Th17. Cell Pathogen. Cell 163, 1413–1427 (2015).
Santori, F. R. et al. Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell Metab. 21, 286–298 (2015).
Orth, J. D., Thiele, I. & Palsson, B. O. What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010).
Chitraju, C. et al. Triglyceride synthesis by DGAT1 protects adipocytes from lipid-induced ER stress during lipolysis. Cell Metab. 26, 407–418 e403 (2017).
Dozsa, A. et al. PPARgamma-mediated and arachidonic acid-dependent signaling is involved in differentiation and lipid production of human sebocytes. J. Invest. Dermatol. 134, 910–920 (2014).
Rudloff, I., Bachmann, M., Pfeilschifter, J. & Muhl, H. Mechanisms of rapid induction of interleukin-22 in activated T cells and its modulation by cyclosporin a. J. Biol. Chem. 287, 4531–4543 (2012).
Bruno, N. E. et al. Creb coactivators direct anabolic responses and enhance performance of skeletal muscle. EMBO J. 33, 1027–1043 (2014).
Riccio, A., Ahn, S., Davenport, C. M., Blendy, J. A. & Ginty, D. D. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science 286, 2358–2361 (1999).
Berod, L. et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014).
Chitraju, C., Walther, T. C. & Farese, R. V. Jr. The triglyceride synthesis enzymes DGAT1 and DGAT2 have distinct and overlapping functions in adipocytes. J. Lipid Res. 60, 1112–1120 (2019).
Nguyen, T. B. et al. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 42, 9–21 e25 (2017).
O’Connor, W. Jr. et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat. Immunol. 10, 603–609 (2009).
Mogi, C. et al. Involvement of proton-sensing TDAG8 in extracellular acidification-induced inhibition of proinflammatory cytokine production in peritoneal macrophages. J. Immunol. 182, 3243–3251 (2009).
Hipolito, V. E. B. et al. Enhanced translation expands the endo-lysosome size and promotes antigen presentation during phagocyte activation. PLoS Biol. 17, e3000535 (2019).
Mindell, J. A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74, 69–86 (2012).
Yambire, K. F. et al. Impaired lysosomal acidification triggers iron deficiency and inflammation in vivo. eLife 8, e51031 (2019).
Alloatti, A. et al. Toll-like receptor 4 engagement on dendritic cells restrains phago-lysosome fusion and promotes cross-presentation of antigens. Immunity 43, 1087–1100 (2015).
Xia, Y. et al. The mevalonate pathway is a druggable target for vaccine adjuvant discovery. Cell 175, 1059–1073.e1021 (2018).
Graham, D. B. et al. ITAM signaling in dendritic cells controls T helper cell priming by regulating MHC class II recycling. Blood 116, 3208–3218 (2010).
Buckley, C. D., Gilroy, D. W., Serhan, C. N., Stockinger, B. & Tak, P. P. The resolution of inflammation. Nat. Rev. Immunol. 13, 59–66 (2013).
Gagliani, N. et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 523, 221–225 (2015).
Schett, G. & Neurath, M. F. Resolution of chronic inflammatory disease: universal and tissue-specific concepts. Nat. Commun. 9, 3261 (2018).
Cartwright, I. M. et al. Adaptation to inflammatory acidity through neutrophil-derived adenosine regulation of SLC26A3. Mucosal Immunol. 13, 230–244 (2020).
Hua, S. Advances in oral drug delivery for regional targeting in the gastrointestinal tract—influence of physiological, pathophysiological and pharmaceutical factors. Front. Pharm. 11, 524 (2020).
Nugent, S. G., Kumar, D., Rampton, D. S. & Evans, D. F. Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut 48, 571–577 (2001).
Gaublomme, J. T. et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 163, 1400–1412 (2015).
Miyazaki, M., Dobrzyn, A., Elias, P. M. & Ntambi, J. M. Stearoyl-CoA desaturase-2 gene expression is required for lipid synthesis during early skin and liver development. Proc. Natl Acad. Sci. USA 102, 12501–12506 (2005).
van Dierendonck, X. et al. HILPDA uncouples lipid droplet accumulation in adipose tissue macrophages from inflammation and metabolic dysregulation. Cell Rep. 30, 1811–1822.e1816 2020).
Geltink, R. I. K., Kyle, R. L. & Pearce, E. L. Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 36, 461–488 (2018).
Suzuki, H., Kumagai, T., Goto, A. & Sugiura, T. Increase in intracellular hydrogen peroxide and upregulation of a nuclear respiratory gene evoked by impairment of mitochondrial electron transfer in human cells. Biochem. Biophys. Res. Commun. 249, 542–545 (1998).
Havlickova Karbanova, V. et al. Compensatory upregulation of respiratory chain complexes III and IV in isolated deficiency of ATP synthase due to TMEM70 mutation. Biochim. Biophys. Acta 1817, 1037–1043 (2012).
Howie, D. et al. A novel role for triglyceride metabolism in Foxp3 expression. Front. Immunol. 10, 1860 (2019).
Gutierrez-Vazquez, C. & Quintana, F. J. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity 48, 19–33 (2018).
Delamarre, L., Pack, M., Chang, H., Mellman, I. & Trombetta, E. S. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307, 1630–1634 (2005).
Trost, M. et al. The phagosomal proteome in interferon-gamma-activated macrophages. Immunity 30, 143–154 (2009).
Graham, D. B. et al. Antigen discovery and specification of immunodominance hierarchies for MHCII-restricted epitopes. Nat. Med. 24, 1762–1772 (2018).
Fleming, S. J., Marioni, J. C. & Babadi, M. CellBender remove-background: a deep generative model for unsupervised removal of background noise from scRNA-seq datasets. Preprint at bioRxiv 791699 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Wagner, A. et al. Metabolic modeling of single Th17 cells reveals regulators of autoimmunity. Cell 184, 4168–4185.e4121 2021).
Seki, A. & Rutz, S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 215, 985–997 (2018).
Acknowledgements
We thank T. Reimels for assistance with editing. This work was supported by funding from the National Institutes of Health (grant nos. R01 DK117263, RC2 DK114784, U19 AI109725 and U19 AI142784 to R.J.X.) and the Helmsley Charitable Trust (to R.J.X.).
Author information
Authors and Affiliations
Contributions
R.J.X. and X.C. conceived of the study. X.C. designed and conducted most of the experiments. A.J. analyzed the scRNA-seq data. Z.C. performed the lipidomic and metabolomic experiments. P.H. and E.A.C. helped with the mouse experiments. N.O.-R. helped with immunoblotting. M.J.D. helped with genetics. X.C. wrote the manuscript. A.J., M.J.D., K.L.C., D.B.G. and R.J.X. revised the manuscript. K.L.C., D.B.G. and R.J.X. supervised the research.
Corresponding author
Ethics declarations
Competing interests
R.J.X. is co-founder of Jnana Therapeutics and Celsius Therapeutics and M.J.D. is a scientific founder of Maze Therapeutics. These organizations played no role in the present study. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 GPR65 I231L mice generated by CRISPR-Cas9 have no evident colitis-related phenotypes at steady state.
(a) Design of GPR65 I231L knock-in by the CRISPR-Cas9 system. Gpr65 gene sequence from NCBI database is shown with annotated sgRNA target sequences and protospacer adjacent motif (PAM) and mutation sites. Mutated alleles are highlighted. (b) Alignment of the sequencing data of WT and I231L mice. Mutated alleles are highlighted. (c) Gating strategy of immune populations in the small intestinal lamina propria. (d,e) Statistical analysis of immune populations in the small intestinal lamina propria (SI LP) and colonic lamina propria (Co LP) of WT (n = 3) and I231L (n = 3) mice. Data are mean values. P values determined by unpaired two-tailed t-test; ns, not significant.
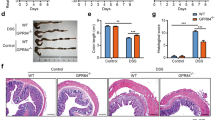
Extended Data Fig. 2 GPR65 I231L mice are more susceptible to bacterial infection-induced colitis.
(a,b) Colony forming units (CFU) in the stool (a) and body weight changes (b) of mice during C. rodentium infection. nWT = 6; nI231L = 7; nKO = 6 mice. (c) H&E-stained sections of distal colon from mice on day 3 after Citrobacter infection. Representative images from one of two independent experiments. Scale bar, 0.5 mm. (d) Statistical analysis of myeloid and CD4+ T cells in the colonic LP on day 12 after C. rodentium infection. nWT = 6; nI231L = 7; nKO = 6 mice. (e) Cytokine-producing CD4+ T cells in the mLN from WT (n = 6), I231L (n = 6) and KO (n = 6) mice on day 12 after infection. (f,g) Cytokine profiles of colonic tissue from C. rodentium-infected mice detected by qPCR (f) and multiplex bead-based cytokine assay (g). n = 4 mice per genotype (f); nWT = 6; nI231L = 7; nKO = 6 mice (g). (h-j) CFU, body weight change and colonic H&E histology of WT (n = 7), I231L (n = 7) or KO (n = 7) CD4+ T cell-transferred mice after Citrobacter infection. CFUs in colonic tissue and H&E staining were performed on day 12. (k,l) Immunophenotyping of CD4+ T cells and inflammatory innate immune cells in the mLN or colonic LP on day 12 after infection. n = 7 mice per genotype. (m) Cytokine profiles of colonic tissue on day 12 were detected by qPCR. n = 4 mice per genotype. Scale bar, 0.5 mm (c,j). Data represent at least two independent experiments. Data are mean values (a,b,d-i,k-m) + SEM (a,b,h,i). P values determined by unpaired two-tailed t-test; ns, not significant.
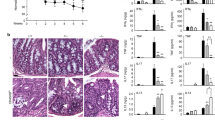
Extended Data Fig. 3 GPR65 regulates Th17 differentiation.
(a) Flow profiles of cell viability of CD4+ T cells after 48 h and 72 h of ex vivo culturing without stimulation. (b) Flow profiles of divided CD4+ T cells after 48 h and 72 h of stimulation with anti-CD3/CD28 dynabeads. (c) In vitro polarization of different Th cells (Th1, Th2, non-pathogenic Th17, pathogenic Th17) and induced Tregs. (d) Cytokine responses to different pH stimulation in Th22 cells polarized in vitro. Intracellular cytokine staining and qPCR were performed after restimulation of resting day 3-polarized Th22 cells by anti-CD3ε and anti-CD28 antibodies for 24 h. (e) Il17a and Rorc expression in polarized Th17 cells and Il22 and Ahr expression in polarized Th22 cells detected by qPCR. Data are mean values (c-e). n = 4 biological replicates for each group (c-e). P values determined by unpaired two-tailed t-test; ns, not significant. Data represent at least two independent experiments.
Extended Data Fig. 4 Single-cell RNA-seq profiling in in vitro polarized Th17 and Th22 cells.
(a,b) UMAP embeddings of single-cell RNA-sequencing profiles from in vitro polarized Th17 and Th22 cells (a). Expression of differential genes across cells reveals the features of different clusters (b). (c) Enrichment of gene ontology metabolic signature scores in cluster 2 single-cell transcriptomes for Th17 and Th22 cells. (d) Flux balance analysis to predict activity of various metabolic processes. Dots denote single biochemical reactions in different metabolisms, and only core biochemical reactions are shown.
Extended Data Fig. 5 Differential expression of genes related to metabolic pathways.
(a) Volcano plots show differential expression of genes related to oxidative phosphorylation (HALLMARK_OXIDATIVE_PHOSPHORYLATION), glycolysis (HALLMARK_GLYCOLYSIS), ATP metabolic process (GO_ATP_METABOLIC_PROCESS) and cellular amide metabolic process (GO_CELLULAR_AMIDE_METABOLIC_PROCESS) in polarized Th17 and Th22 cells (KO versus WT). (b) Dot plot shows the differential expression of genes related to different metabolic processes in all single-cell transcriptomes. (c) qPCR validation of differential gene expression in I231L Th17 and Th22 cells. Data are mean values from two independent experiments. n = 4 biological replicates for each group. P values determined by unpaired two-tailed t-test; ns, not significant.
Extended Data Fig. 6 Metabolomics analysis in polarized Th17 and Th22 cells.
Heatmap shows all 160 lipid metabolites detected in lipidomics.
Extended Data Fig. 7 GPR65 I231L exacerbates T cell-driven colitis.
(a) Body weight changes of Rag1 KO mice after transfer of Gpr65 WT, I231L or KO (n = 6 per genotype) CD45RBhigh CD4+ T cells. (b) Colon length of mice (n = 6 per genotype) in (a) after colitis induction. (c) H&E-stained sections of distal colon from mice with T cell-driven colitis. Representative images from one of two independent experiments. Scale bar, 0.5 mm. (d) Proportion analysis of cytokine-producing CD4+ T cells in the mLN and colonic LP of mice (n = 6 per genotype) in (a) after colitis induction. (e) Cytokine profiles (IFNγ, IL-17A, IL-22) of colonic tissue from mice (n = 4 per genotype) with colitis detected by multiplex bead-based cytokine assay. (f) Cytokine profiles (IFNγ, IL-17A, IL-22) of colonic tissue from mice (n = 4 per genotype) with T cell-driven colitis detected by qPCR. One dot denotes one biological replicate (b,d-f). (g) Gating strategy of immune populations in the colonic LP. Data are mean values (a,b,d-f) + SEM (a). P values determined by unpaired two-tailed t-test; ns, not significant. Data represent at least two independent experiments.
Extended Data Fig. 8 GPR65 I231L enhances antigen presentation to CD4+ T cells by dendritic cells.
(a,b) BMDC:OT-II T cell co-culturing-based antigen presentation assay. Cell numbers of OT-II CD4+ T cells (a) and IL-2 cytokine in the culture supernatant (b) on day 3 after co-culturing are shown. n = 5 (WT), n = 3 (I231L) and n = 2 (KO) biological replicates (a); n = 4 for each group (b). (c,d) BMDCs incubated with both DQ-Red BSA and AF647-BSA with or without treatment. Cells were treated with LPS (20 ng/ml) for 2 h and then incubated with DQ-Red BSA (3 μg/ml) and AF647-BSA (3 μg/ml) for 1 h before imaging. Representative images from one of two independent experiments are shown (c). Scale bar, 10μm. Statistical analysis of the ratio of DQ-Red BSA and AF647 fluorescence intensity (d). nWT = 39, nI231L = 28, nKO = 24 (NT); nWT = 27, nI231L = 20, nKO = 20 (LPS); nWT = 26, nI231L = 19, nKO = 20 (LPS + pH6.8); nWT = 21, nI231L = 18, nKO = 14 (Baf A1) cells. Data are mean values (a,b,d) + SEM (a,b). P values determined by unpaired two-tailed t-test; ns, not significant. Data represent at least two independent experiments.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Chen, X., Jaiswal, A., Costliow, Z. et al. pH sensing controls tissue inflammation by modulating cellular metabolism and endo-lysosomal function of immune cells. Nat Immunol 23, 1063–1075 (2022). https://doi.org/10.1038/s41590-022-01231-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-022-01231-0
This article is cited by
-
What role does GPR65 play in the progression of osteosarcoma? Its mechanism and clinical significance
Cancer Cell International (2024)
-
Role of pH-sensing receptors in colitis
Pflügers Archiv - European Journal of Physiology (2024)
-
Recent advances in acid sensing by G protein coupled receptors
Pflügers Archiv - European Journal of Physiology (2024)
-
Basic amino acid-mediated cationic amphiphilic surfaces for antimicrobial pH monitoring sensor with wound healing effects
Biomaterials Research (2023)
-
A molecular characterization and clinical relevance of microglia-like cells derived from patients with panic disorder
Translational Psychiatry (2023)