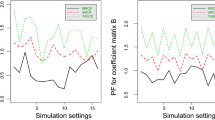
Abstract
The paper addresses joint sparsity selection in the regression coefficient matrix and the error precision (inverse covariance) matrix for high-dimensional multivariate regression models in the Bayesian paradigm. The selected sparsity patterns are crucial to help understand the network of relationships between the predictor and response variables, as well as the conditional relationships among the latter. While Bayesian methods have the advantage of providing natural uncertainty quantification through posterior inclusion probabilities and credible intervals, current Bayesian approaches either restrict to specific sub-classes of sparsity patterns and/or are not scalable to settings with hundreds of responses and predictors. Bayesian approaches that only focus on estimating the posterior mode are scalable, but do not generate samples from the posterior distribution for uncertainty quantification. Using a bi-convex regression-based generalized likelihood and spike-and-slab priors, we develop an algorithm called joint regression network selector (JRNS) for joint regression and covariance selection, which (a) can accommodate general sparsity patterns, (b) provides posterior samples for uncertainty quantification, and (c) is scalable and orders of magnitude faster than the state-of-the-art Bayesian approaches providing uncertainty quantification. We demonstrate the statistical and computational efficacy of the proposed approach on synthetic data and through the analysis of selected cancer data sets. We also establish high-dimensional posterior consistency for one of the developed algorithms.








Similar content being viewed by others
References
Alquier, P.: Approximate bayesian inference. Entropy (Basel) 22, 1272 (2020)
Barbieri, M.M., Berger, J.O.: Optimal predictive model selection. Ann. Stat. 32(3), 870–897 (2004)
Besag, J.: Statistical analysis of non-lattice data. J. R. Stat. Soc.: Ser. D (The Statistician) 24(3), 179–195 (1975)
Bhadra, A., Mallick, B.K.: Joint high-dimensional Bayesian variable and covariance selection with an application to eQTL analysis. Biometrics 69(2), 447–457 (2013)
Bissiri, P., Holmes, C., Walker, S.: A general framework for updating belief distributions. J. R. Stat. Soc. Ser. B 78, 1103–1130 (2016)
Brown, P.J., Vannucci, M., Fearn, T.: Multivariate Bayesian variable selection and prediction. J. R. Stat. Soc. Ser. B 60, 627–641 (1998)
Cai, T.T., Li, H., Liu, W., Xie, J.: Covariate-adjusted precision matrix estimation with an application in genetical genomics. Biometrika 100(1), 139–156 (2013)
Cao, X., Khare, K., Ghosh, M.: Posterior graph selection and estimation consistency for high-dimensional bayesian dag models. Ann. Stat. 47(1), 319–348 (2019)
Consonni, G., La Rocca, L., Peluso, S.: Objective Bayes covariate-adjusted sparse graphical model selection. Scand. J. Stat. 44, 741–764 (2017)
Deshpande, S.K., Ročková, V., George, E.I.: Simultaneous variable and covariance selection with the multivariate spike-and-slab lasso. J. Comput. Gr. Stat. 28(4), 921–931 (2019)
Friedman, J., Hastie, T., Tibshirani, R.: Sparse inverse covariance estimation with the graphical lasso. Biostatistics 9(3), 432–441 (2008)
Van de Geer, S., Bühlmann, P., Ritov, Y., Dezeure, R.: On asymptotically optimal confidence regions and tests for high-dimensional models. Ann. Stat. 42(3), 1166–1202 (2014)
Gonzalez, D.M., Medici, D.: Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 7(344), 8 (2014)
Ha, MJ., Stingo, F., Baladandayuthapani, V.: Supplemental material for ‘Bayesian Structure Learning in Multi-layered Genomic Networks’. Github (2020a)
Ha, M.J., Stingo, F.C., Baladandayuthapani, V.: Bayesian structure learning in multi-layered genomic networks. J. Am. Stat. Assoc. 1, 1–33 (2020b)
Khare, K., Oh, S., Rajaratnam, B.: A convex pseudolikelihood framework for high dimensional partial correlation estimation with convergence guarantees. J. R. Stat. Soc. B 77, 803–825 (2015)
Lee, K., Lee, K., Lee, J.: Post-processed posteriors for banded covariances. arXiv preprint arXiv:2011.12627 (2020)
Lee, W., Liu, Y.: Simultaneous multiple response regression and inverse covariance matrix estimation via penalized gaussian maximum likelihood. J. Multivar. Anal. 111, 241–255 (2012)
Li, Y., Datta, J., Craig, B.A., Bhadra, A.: Joint mean-covariance estimation via the horseshoe. J. Multivar. Anal. 183, 104716 (2021)
Lin, J., Basu, S., Banerjee, M., Michailidis, G.: Penalized maximum likelihood estimation of multi-layered gaussian graphical models. J. Mach. Learn. Res. 17, 1–51 (2016)
Lin, L., Drton, M., Shojaie, A.: High-dimensional inference of graphical models using regularized score matching. Electron. J. Stat. 10(1), 394–422 (2016)
Ma, J., Michailidis, G.: Joint structural estimation of multiple graphical models. J. Mach. Learn. Res. 17(166), 1–48 (2016)
McCarter, C., Kim, S.: (2014) On sparse gaussian chain graph models. In: Advances in Neural Information Processing Systems, pp. 3212–3220
Meinshausen, N., Bühlmann, P.: High-dimensional graphs and variable selection with the lasso. Ann. Stat. 1, 1436–1462 (2006)
Narisetty, N., He, X.: Bayesian variable selection with shrinking and diffusing priors. Ann. Stat. 42, 789–817 (2014)
Peng, J., Wang, P., Zhou, N., Zhu, J.: Partial correlation estimation by joint sparse regression models. J. Am. Stat. Assoc. 104, 735–746 (2009)
Richardson, S., Bottolo, L., Rosenthal, J.S.: Bayesian models for sparse regression analysis of high dimensional data. In: Bernardo, J., Bayarri, M., Berger, J., Dawid, A., Heckerman, D., Smith, A.F.M., West, M. (Eds.) Bayesian Statistics 9 (2010)
Rothman, A.J., Levina, E., Zhu, J.: Sparse multivariate regression with covariance estimation. J. Comput. Gr. Stat. 19(4), 947–962 (2010)
Sohn, KA., Kim, S.: (2012) Joint estimation of structured sparsity and output structure in multiple-output regression via inverse-covariance regularization. In: International Conference on Artificial Intelligence and Statistics, pp 1081–1089
Wang, H.: Bayesian graphical lasso models and efficient posterior computation. Bayesian Anal. 7(4), 867–886 (2012)
Yuan, X.T., Zhang, T.: Partial gaussian graphical model estimation. IEEE Trans. Inf. Theory 60(3), 1673–1687 (2014)
Acknowledgements
The work of GM was supported in part by NIH grants 1U01CA235489-01 and 1R01GM114029-01A1.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Appendix: Pathways for TCGA cancer data
Appendix: Pathways for TCGA cancer data
Table 13 lists the indices of all the genes and proteins in the LUAD cancer data, and Table 14 lists all the pathways that have been considered in the analysis of the TCGA cancer data in Sect. 5 and their gene members.
Rights and permissions
About this article
Cite this article
Samanta, S., Khare, K. & Michailidis, G. A generalized likelihood-based Bayesian approach for scalable joint regression and covariance selection in high dimensions. Stat Comput 32, 47 (2022). https://doi.org/10.1007/s11222-022-10102-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11222-022-10102-5