Abstract
Impaired chronic viral and tumor clearance has been attributed to CD8+ T cell exhaustion, a differentiation state in which T cells have reduced and altered effector function that can be partially reversed upon blockade of inhibitory receptors. The role of the exhaustion program and transcriptional networks that control CD8+ T cell function and fate in autoimmunity is not clear. Here we show that intra-islet CD8+ T cells phenotypically, transcriptionally, epigenetically and metabolically possess features of canonically exhausted T cells, yet maintain important differences. This ‘restrained’ phenotype can be perturbed and disease accelerated by CD8+ T cell-restricted deletion of the inhibitory receptor lymphocyte activating gene 3 (LAG3). Mechanistically, LAG3-deficient CD8+ T cells have enhanced effector-like functions, trafficking to the islets, and have a diminished exhausted phenotype, highlighting a physiological role for an exhaustion program in limiting autoimmunity and implicating LAG3 as a target for autoimmune therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The following databases were used for analysis (Methods). Mouse genome for alignment of sequencing data: Genome Reference Consortium Mouse Build 38 (Figs. 1b and 2, Extended Data Figs. 2c,d and 5–7; https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/); HINT—to generate PPI networks (Extended Data Fig. 8http://hint.yulab.org/)51, KEGG (Extended Data Fig. 5ehttps://www.genome.jp/kegg/)44,45,46 and C7 Immunologic Signature Gene Sets (Extended Data Fig. 5e; http://www.gsea-msigdb.org/gsea/msigdb/collections.jsp)82.
The following datasets were used for analysis: ATAC–seq GSEA (Fig. 1c and Extended Data Fig. 2e;GSE86797; ref. 70), bulk RNA-seq GSEA (Extended Data Fig. 2c; GSE41867; ref. 75) and scRNA-seq GSEA (Fig. 3c, GSE122713; ref. 26).
Sequencing data generated in this manuscript are deposited in the Gene Expression Omnibus database under accession code GSE199357. Source data are provided with this paper.
Code availability
Standard R packages were used for data analysis and generation of figures as described in the Methods. Code for previously described custom packages is available at www.github.com/arc85/dragonsc/. Specific code to generate figures is available upon request.
References
Blank, C. U. et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 19, 665–674 (2019).
Beltra, J. C. et al. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52, 825–841 (2020).
Khan, O. et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019).
Scott, A. C. et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019).
Seo, H. et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl Acad. Sci. USA 116, 12410–12415 (2019).
Yao, C. et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20, 890–901 (2019).
Tilstra, J. S. et al. Kidney-infiltrating T cells in murine lupus nephritis are metabolically and functionally exhausted. J. Clin. Invest. 128, 4884–4897 (2018).
Smita, S., Chikina, M., Shlomchik, M. J. & Tilstra, J. S. Heterogeneity and clonality of kidney-infiltrating T cells in murine lupus nephritis. JCI Insight https://doi.org/10.1172/jci.insight.156048 (2022).
Long, S. A. et al. Remodeling T cell compartments during anti-CD3 immunotherapy of type 1 diabetes. Cell Immunol. 319, 3–9 (2017).
McKinney, E. F., Lee, J. C., Jayne, D. R., Lyons, P. A. & Smith, K. G. T cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616 (2015).
Wiedeman, A. E. et al. Autoreactive CD8+ T cell exhaustion distinguishes subjects with slow type 1 diabetes progression. J. Clin. Invest. 130, 480–490 (2020).
Long, S. A. et al. Partial exhaustion of CD8+ T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci. Immunol. https://doi.org/10.1126/sciimmunol.aai7793 (2016).
Pauken, K. E., Dougan, M., Rose, N. R., Lichtman, A. H. & Sharpe, A. H. Adverse events following cancer immunotherapy: obstacles and opportunities. Trends Immunol. 40, 511–523 (2019).
Burke, K. P., Grebinoski, S., Sharpe, A. H. & Vignali, D. A. A. Understanding adverse events of immunotherapy: a mechanistic perspective. J. Exp. Med. https://doi.org/10.1084/jem.20192179 (2020).
Gearty, S. V. et al. An autoimmune stem-like CD8+ T cell population drives type 1 diabetes. Nature https://doi.org/10.1038/s41586-021-04248-x (2021).
Chen, Y. -G., Mathews, C. E. & Driver, J. P. The role of NOD mice in type 1 diabetes research: lessons from the past and recommendations for the future. Front. Endocrinol. https://doi.org/10.3389/fendo.2018.00051 (2018).
Christianson, S. W., Shultz, L. D. & Leiter, E. H. Adoptive transfer of diabetes into immunodeficient NOD-scid/scid mice. Relative contributions of CD4+ and CD8+ T cells from diabetic versus prediabetic NOD.NON-Thy-1a donors. Diabetes 42, 44–55 (1993).
Varela-Calvino, R., Calviño-Sampedro, C., Gómez-Touriño, I. & Cordero, O. J. Apportioning blame: autoreactive CD4+ and CD8+ T cells in type 1 diabetes. Arch. Immunol. Ther. Exp. 65, 275–284 (2017).
Yeo, L. et al. Autoreactive T effector memory differentiation mirrors β cell function in type 1 diabetes. J. Clin. Invest. 128, 3460–3474 (2018).
Skowera, A. et al. β-cell-specific CD8+ T cell phenotype in type 1 diabetes reflects chronic autoantigen exposure. Diabetes 64, 916–925 (2015).
Abdelsamed, H. A. et al. Beta cell-specific CD8+ T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes. Nat. Immunol. 21, 578–587 (2020).
Zakharov, P. N., Hu, H., Wan, X. & Unanue, E. R. Single-cell RNA sequencing of murine islets shows high cellular complexity at all stages of autoimmune diabetes. J. Exp. Med. https://doi.org/10.1084/jem.20192362 (2020).
Bettini, M. et al. Cutting edge: accelerated autoimmune diabetes in the absence of LAG-3. J. Immunol. 187, 3493–3498 (2011).
Kotecha, N., Krutzik, P. O. & Irish, J. M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 10, 17 (2010).
Zhang, Q. et al. LAG3 limits regulatory T cell proliferation and function in autoimmune diabetes. Sci. Immunol. https://doi.org/10.1126/sciimmunol.aah4569 (2017).
Miller, B. C. et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Chee, J. et al. Effector-memory T cells develop in islets and report islet pathology in type 1 diabetes. J. Immunol. 192, 572–580 (2014).
Kuric, E. et al. Demonstration of tissue-resident memory CD8+ T cells in insulitic lesions in adult patients with recent-onset type 1 diabetes. Am. J. Pathol. 187, 581–588 (2017).
Lennon, G. P. et al. T cell islet accumulation in type 1 diabetes is a tightly regulated, cell-autonomous event. Immunity 31, 643–653 (2009).
Paley, M. A. et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012).
Li, J., He, Y., Hao, J., Ni, L. & Dong, C. High levels of Eomes promote exhaustion of anti-tumor CD8+ T cells. Front. Immunol. 9, 2981 (2018).
Seo, W., Jerin, C. & Nishikawa, H. Transcriptional regulatory network for the establishment of CD8+ T cell exhaustion. Exp. Mol. Med. 53, 202–209 (2021).
Canale, F. P. et al. CD39 expression defines cell exhaustion in tumor-infiltrating CD8+ T cells. Cancer Res. 78, 115–128 (2018).
Gupta, P. K. et al. CD39 expression identifies terminally exhausted CD8+ T cells. PLoS Pathog. 11, e1005177 (2015).
Bengsch, B. et al. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8+ T cell exhaustion. Immunity 45, 358–373 (2016).
Scharping, N. E. et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 22, 205–215 (2021).
Scharping, N. E. et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 701–703 (2016).
Sato, Y. et al. Cellular hypoxia of pancreatic beta-cells due to high levels of oxygen consumption for insulin secretion in vitro. J. Biol. Chem. 286, 12524–12532 (2011).
Olsson, R., Olerud, J., Pettersson, U. & Carlsson, P.-O. Increased numbers of low-oxygenated pancreatic islets after intraportal islet transplantation. Diabetes 60, 2350–2353 (2011).
Olsson, R. & Carlsson, P. O. A low-oxygenated subpopulation of pancreatic islets constitutes a functional reserve of endocrine cells. Diabetes 60, 2068–2075 (2011).
Komatsu, H., Kandeel, F. & Mullen, Y. Impact of oxygen on pancreatic islet survival. Pancreas 47, 533–543 (2018).
Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z. & Zhang, B. WebGestalt 2019: gene-set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–w205 (2019).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K. & Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–d595 (2019).
Priatel, J., Huang, Y.-H., Tsai, K., Harder, K. & Tan, R. SLAM-associated protein modulates CD8+ T cell responses primed by antigen-presenting B cells (IRM4P.503). J. Immunol. 192, 61.10 (2014).
Creelan, B. C. & Antonia, S. J. The NKG2A immune checkpoint—a new direction in cancer immunotherapy. Nat. Rev. Clin. Oncol. 16, 277–278 (2019).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Haghverdi, L., Büttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016).
Das, J. & Yu, H. HINT: High-quality protein interactomes and their applications in understanding human disease. BMC Syst. Biol. 6, 92 (2012).
Cusick, M. E. et al. Literature-curated protein interaction datasets. Nat. Methods 6, 39–46 (2009).
Kent, S. C. et al. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature 435, 224–228 (2005).
Nakayama, M. et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 435, 220–223 (2005).
Krishnamurthy, B. et al. Responses against islet antigens in NOD mice are prevented by tolerance to proinsulin but not IGRP. J. Clin. Invest. 116, 3258–3265 (2006).
Prasad, S., Xu, D. & Miller, S. D. Tolerance strategies employing antigen-coupled apoptotic cells and carboxylated PLG nanoparticles for the treatment of type 1 diabetes. Rev. Diabet. Stud. 9, 319–327 (2012).
Amrani, A. et al. Progression of autoimmune diabetes driven by avidity maturation of a T cell population. Nature 406, 739–742 (2000).
Woo, S. R. et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T cell function to promote tumoral immune escape. Cancer Res. 72, 917–927 (2012).
Andrews, L. P. et al. Resistance to PD1 blockade in the absence of metalloprotease-mediated LAG3 shedding. Sci. Immunol. https://doi.org/10.1126/sciimmunol.abc2728 (2020).
Blackburn, S. D. et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10, 29–37 (2009).
Grebinoski, S. & Vignali, D. A. A. Inhibitory receptor agonists: the future of autoimmune disease therapeutics? Curr. Opin. Immunol. 67, 1–9 (2020).
Jones, B. E. et al. Fewer LAG-3+ T cells in relapsing–remitting multiple sclerosis and type 1 diabetes. J. Immunol. https://doi.org/10.4049/jimmunol.2100850 (2022).
Leiter, E. H. The NOD mouse: a model for insulin-dependent diabetes mellitus. Curr. Protoc. Immunol. 15, 15.19 (2001).
Rodriguez-Calvo, T., Ekwall, O., Amirian, N., Zapardiel-Gonzalo, J. & von Herrath, M. G. Increased immune cell infiltration of the exocrine pancreas: a possible contribution to the pathogenesis of type 1 diabetes. Diabetes 63, 3880–3890 (2014).
Watson, M. J. et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 591, 645–651 (2021).
Roederer, M., Nozzi, J. L. & Nason, M. C. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 79, 167–174 (2011).
Pauken, K. E. et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 3 (2004).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene-set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Doering, T. A. et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37, 1130–1144 (2012).
Stoeckius, M. et al. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Cillo, A. R. et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity 52, 183–199 (2020).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2009).
Wickham, H. et al. Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 (2019).
Godec, J. et al. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity 44, 194–206 (2016).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
We thank the current and former members in the Vignali laboratory (Vignali-lab.com; @Vignali_Lab) for their constructive comments and advice during this project. The authors also thank the members of P01 AI108545 for their constructive feedback, specifically A. Sharpe and laboratory members (Department of Microbiology and Immunobiology, Harvard Medical School). We thank G. Delgoffe and laboratory members for aid in metabolic studies and protocols (Department of Immunology, University of Pittsburgh; Tumor Microenvironment Center, UPMC Hillman Cancer Center; Cancer Immunology and Immunotherapy Program, UPMC Hillman Cancer Center). We also thank Q. Zhan for help with histology (Department of Pathology, Brigham and Women’s Hospital and Harvard Medical School). We thank R. Dadey, H. Yano, L. Andrews, R. Peralta and P. Vignali, members of the Vignali and Delgoffe laboratories, for aid in unpublished experiments. We would also like to thank Clayton Mathews (Department of Pathology, University of Florida) and members of his lab Jing Chen, Sadie Clements, and Mollie Huber for aid and reagents for unpublished experiments. We thank D. Falkner, A. McIntyre, N. Sheng, H. Gunzelman, T. Sturgeon, H. Shen and L. Borghesi from the Immunology Flow Core (current and past) for cell sorting, maintenance and training in the use of cytometers (Department of Immunology, UPSOM). We thank J. Goulet, R. Gerheart, C. Schmidt, J. Latini and the staff of the Division of Laboratory Animal Resources for genotyping and animal husbandry (Department of Immunology, UPSOM). We thank the Littman (The Kimmel Center for Biology and Medicine of the Skirball Institute, New York University School of Medicine; Howard Hughes Medical Institute) and Taniuchi (Laboratory for Transcriptional Regulation, RIKEN Center for Integrative Medical Sciences) laboratories for the E8iCRE-GFP.C57BL/6 mice and A. Castellaw for breeding them onto the NOD background at St. Jude Children’s Research Hospital (Department of Immunology). We also thank the UPMC Genome Core and the University of Pittsburgh HSCRF Genomics Research Core for assistance with next-generation sequencing, and the Pitt Center for Research Computing for computational resources. This work was supported by the NIH (R01 DK089125, R01 AI144422, P01 AI108545 to D.A.A.V.; AI105343, AI117950, AI082630, AI112521, AI115712, AI108545 and CA210944 to E.J.W.; DP2AI164325 and U01HG012041 to J.D. in partial support of his contributions; F31 AI147638 and T32 AI089443 to S.G.). E.J.W. is supported by the Parker Institute for Cancer Immunotherapy, which supports the cancer immunology program at UPenn. A.R.C. is supported by the Hillman Postdoctoral Fellows for Innovative Cancer Research postdoctoral fellowship. This project benefitted from a Special Order BD LSRFortessa (funded by NIH S10 OD011925) used in the UPSOM Unified Flow Core.
Author information
Authors and Affiliations
Contributions
D.A.A.V. conceived, directed and obtained funding for the project. S.G., Q.Z., C.J.W. and D.A.A.V. conceptualized, designed and analyzed the experiments. S.G. and D.A.A.V. wrote the manuscript with input from J.D., H.X., S.M., C.J.W. and A.R.C. S.G. and Q.Z. performed all experiments. A.R.C. and T.T. helped conceptualize scRNA-seq experiments and A.R.C. and S.M. analyzed the scRNA-seq and TCR-seq data. S.M. analyzed the bulk RNA-seq data. J.D. and H.X. synthesized protein interaction networks and PLS-DA. E.A.B. aided in diabetic mouse colony maintenance, breeding and diabetes incidence. T.T. and C.C. generated single-cell libraries. C.G.L. and G.F.M. oversaw H&E staining. R.L. contributed advice for RNA-seq library generation and data acquisition. E.J.W. contributed advice in experimental design and analysis. J.D. and H.X. contributed to RNA-seq analysis, network analysis and interpretation. C.J.W. contributed to experimental design, analysis and developing mouse strains. All authors provided feedback and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.A.A.V. and C.J.W. declare competing financial interests and have submitted patents covering LAG3 that are licensed or pending and are entitled to a share in net income generated from licensing of these patent rights for commercial development. D.A.A.V. is cofounder and stock holder of Novasenta, Potenza, Tizona and Trishula; stock holder of Oncorus, Werewolf and Apeximmune; has patents licensed and royalties from Astellas, BMS, Novasenta; scientific advisory board member of Tizona, Werewolf, F-Star, Bicara, Apeximmune and T7/Imreg Bio; is a consultant for Astellas, BMS, Almirall, Incyte, G1 Therapeutics and Inzen Therapeutics; and obtained research funding from BMS, Astellas and Novasenta. E.J.W. has consulting agreements with and/or is on the scientific advisory board for Merck, Roche, Pieris, Elstar and Surface Oncology. E.J.W. has a patent licensing agreement on the PD1 pathway with Roche/Genentech. E.J.W. is a founder of Arsenal Biosciences. The other authors declare no competing interests.
Peer review
Peer review information
Primary Handling Editor: N. Bernard, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Intra-Islet CD8+ T cells upregulate markers of exhaustion but are a heterogeneous population.
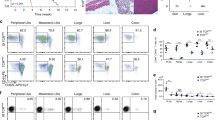
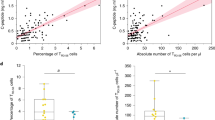
Phenotypic quantification of exhaustion markers in the NOD model of diabetes. (a-f) Spectral flow cytometry for CD8+ T cell functional markers was completed over a time course of 6-14-week-old female WT NOD mice. Representative flow plots are derived from total intra-islet CD8+ T cells (gated on lymphocytes, single cells, Live, Thy1.2+, CD8b+) of 12-week-old female NOD. Data were accumulated from a total of 5 experiments, each experiment had mice of several ages, with n = 10 mice per timepoint, n = 50 total mice. Each point on the graph is representative of a single mouse. (a) Representative flow plotting demonstrating gating strategy to obtain CD8+ T cells. (b) High dimensional analysis at 12 weeks of age was preformed using Cytobank38 viSNE map analysis (Methods). viSNE maps are shown portraying the 11 markers are used to create FlowSOM clustering analysis. (c) Representative flow plots of intra-islet CD8+ T cell PD1 and LAG3 (6 vs 12 weeks p = .0355), TIGIT (6 vs 12, 14 weeks p = .0005, 0.0011), TIM3, CTLA4, and ICOS expression are shown islets and expression of IRs are quantified from the ndLN, pLN, and islets. (d) Co-expression of multiple IRs and the transcription factor TOX are represented in Simplified Presentation of Incredibly Complex Evaluations (SPICE)63 plots showing bulk CD8+ T cells from 6- and 12-week-old islet samples. (e) Representative flow plots and quantification of bulk intra-islet CD8+ T cell expression of TCF1 and TOX populations. (TCF1–TOX + 6 vs 12, 14 weeks, p = 0.0029, 0.0021) (f) % TOX+ correlation to PD1 (p < .0001), TIGIT (p < .0001), and LAG3 (p < .0001). Pearson’s correlation coefficients and r2 values were calculated. (c and e) A two-sided nonparametric Mann-Whitney was preformed. Graphs portray the median. P = * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. Unlabeled indicates not statistically significant.
Extended Data Fig. 2 Intra-islet CD8+ T cells express LAG3, which marks exhausted CD8+ T cells, though total intra-islet CD8+ T cells also share features of effector T cells.
Transcriptional and epigenetic analysis was performed on intra-islet CD8+ T cells (a) WT Lag3 locus is shown in the top panel. The Lag3L/L-YFP construct is generated by inserting LoxP sites flanking the transmembrane region, exon 7, of the Lag3 gene (middle panel). (b) YFP expression is demonstrated in the Lag3L/L-YFP.NOD, marking those CD8+ T cells which have transcribed Lag3. (c-d) Bulk population RNAseq was preformed comparing intra-islet YFP+ and YFP– CD8+ T cells, along with YFP– ndLN and pLN controls. Cells are pooled from 3 Lag3L/L-YFP.NOD 8 week old females in 2 independent experiments. (c) Relative expression of selected co-stimulatory or co-inhibitory receptors in the YFP+ vs YFP– intra-islet CD8+ T cells. (d) Leading-edge gene set enrichment analysis was preformed comparing YFP+ and YFP– intra-islet CD8+ T cells to published exhaustion64 and activation65 datasets. NES = Normalized Enrichment score, fdr = false discovery rate. (Methods) (e) scATACseq was preformed comparing E8iCRE/CRE-GFP.NOD CD8+ T cells derived from islets and ndLN (n = 4, 8 week Females). Enrichment for effector signature peaks is shown.
Extended Data Fig. 3 ~50% of intra-islet CD8+ T cells express markers of memory, while only a small fraction express marker of naivety or effector function, while Tetramer+ cells have minimal changes in phenotype with disease progression.
Flow cytometric quantification of markers associated with naïve, effector, and memory CD8+ T cell subsets. (a-g) Spectral flow cytometry for CD8+ T cell functional markers was completed over a timecourse of 6-14-week-old female WT NOD mice. Representative flow plots are derived from total intra-islet CD8+ T cells (gated on lymphocytes, single cells, Live, Thy1.2+, CD8b+) of 12-week-old female NOD. Data were accumulated from a total of 5 experiments, each experiment had mice of several ages with n = 10 mice per timepoint, n = 50 total mice. Each point on the graph is representative of a single mouse. Data shown is analyzing total intra-islet CD8+ T cells, gated on Live, Thy1.2+, CD8b+ or PD1+ vs PD1– intra-islet CD8+ T cells. (a) Representative flow plot and quantification of CD127 expression on total ndLN, pLN and intra-islet CD8+ T cells (6 vs. 8, 12, 14 weeks, p = .0288, 0.0089, 0.063) (b) Representative flow plot and quantification of CD127 expression on or PD1+ vs PD1– intra-islet CD8+ T cells (PD1+ vs PD1– p < .000001 at all time points, 6 vs 12, 14 weeks PD1+ p = .05, 0.05). (c) Quantification of CD62L in islets compared to ndLN and pLN, as well as on intra-islet PD1+ vs PD1– populations. (d) Representative flow plot and MFI of CD44 expression on ndLN, pLN, islet, and islet PD1 subsets. (e) Representative flow plot of KLRG1 expression and quantification of KLRG1 on ndLN, pLN, islet, and islet PD1 subsets (PD1+ vs PD1– 6, 8, 10, 12, 14 weeks p = .0288, 0.0011, 0.0003, 0.000076, 0.000011, 6 vs 14 weeks PD1+ p = .055). (f) Quantification of tetramer+ CD8+ T cells in the islet’s over time. (g) Expression of CD8+ T cell functional markers on tetramer+ populations in the islets. Only samples consisting of >40 Tetramer+ CD8+ T cells are shown. Tetramer staining in lymph nodes was negligible and never exceeded 40 tetramer+ cells. (a-g) Each data point corresponds to a single mouse. A two-sided nonparametric Mann-Whitney was preformed, where P = * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. Unlabeled indicates not statistically significant. Graphs portray the median.
Extended Data Fig. 4 A subset of intra islet CD8+ T cells upregulate markers of exhaustion, as well as effector cell markers.
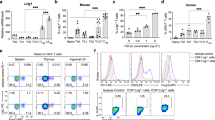
(a-d) Spectral flow cytometry for CD8+ T cell functional markers was completed and representative flow plots and graphs appear as described in Extended Data Fig. 1, with the added sub gate of PD1+ and PD1–. (a) representative flow plot of intra-islet CD8+ T cells PD1 expression. (b) Representative flow plots and quantification of LAG3 and TIGIT expression on PD1+ and PD1– intra-islet CD8+ T cells. (LAG3+TIGIT+: PD1+ vs PD1– 6, 8, 10-14 weeks p = .0005, 0.000002, <.000001, 6 vs 12, 14 weeks PD1+ p = .0039, 0.0065. LAG3–TIGIT–: PD1+ vs PD1– 6, 8-14 weeks p = .000174, <.000001, 6 vs 10, 12, 14 weeks PD1+ p = .028, 0.006, 0.005). (c) Representative flow plots of TCF1 and TOX staining on PD1+ and PD1– intra-islet CD8+ T cells. (d) quantification of (c) (TCF1+TOX–: PD1+ vs PD1– p = .000011 at all time points, 6 vs 10, 12, 14 weeks PD1+ p = .0027, 0.0019, 0.0064. TCF1+TOX+: PD1+ vs PD1– p = <.000001 at all timepoints. TCF1–TOX+: PD1+ vs PD1– 6, 8-14 weeks p = .000262, <.000001, 6 vs 8, 10, 12, 14 weeks p = .0355, 0.0355, 0.0147, 0.0014). (e-f) Total intra-islet CD8+ T cells from 12-week-old female WT NOD mice were analyzed by spectral flow cytometry including ndLN and pLN controls (n = 10, 2 independent experiments). (e) Representative flow plots (islets) and quantification of CD73 and CD39 expression (CD73+: islets vs ndLN, pLN p = <.0001, 0.06. CD39+CD73+: islets vs ndLN, pLN, p = <.0001, 0.0142. CD39+: islets vs ndLN, pLN, p = <.0001, 0.0315). (f) Representative flow plots (islets) and quantification of Tbet and Eomes expression (Tbet+: islets vs ndLN, pLN, p = .0056, 0.0003. Eomes+: islets vs ndLN, pLN, p = .0005, <.0001). (g-i) intra-islet CD8+ T cells were isolated and from 12-week-old female WT NODs and analyzed by flow cytometry for metabolic markers or cytokines (n = 10, 2 independent experiments, techniques described in methods) (g) intra-islet CD8+ T cells are stained for TMRM (islets vs ndLN, pLN p = <.0001, 0.0106), (h) MitoSOX (islets vs. ndLN and pLN, p < .0001) and CellROX (islets vs ndLN p = .0019), and for (i) cytokine production. (a-i) Each data point corresponds to a single mouse. A two-sided nonparametric Mann-Whitney was preformed, where P = * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. Unlabeled indicates not statistically significant. Graphs portray the median.
Extended Data Fig. 5 scRNAseq reveals transcriptionally unique clusters and functions of Cre Control versus Lag3∆TM CD8+ T cells.
scRNAseq assessment of intra-islet CD8+ T cells. (a) The Lag3L/L-YFP (Extended Data Fig. 2a) construct crossed to a Cre recombinase is shown. Upon crossing Lag3L/L-YFP to a Cre recombinase, exon 7 (the transmembrane domain) is deleted (Lag3∆TM). The result is the generation of only the soluble form of LAG3 protein. (b) qPCR determining deletion efficiency of the CD8 specific LAG3ΔTM mouse. Ratio of Exon 7 to Exon 3 was quantified in Cre Control (E8ICRE/CRE-GFP.NOD), vs Lag3∆TM (Lag3L/L-YFPE8ICRE/CRE-GFP.NOD) experimental mice. Cells derived from spleens of five 8-week-old females for 1 experiment (n = 5). (c-g) CD8+ T cells from the islets and ndLN were isolated from 4 Cre Control and 4 Lag3∆TM 8-week-old NOD female mice and were subjected to 5’ paired single cell RNAseq (scRNAseq) and single cell T cell receptor sequencing (scTCRseq). (c) Cells were visualized by UMAP and colored by tissue, genotype, or individual sample. (d) Quantification of specific cell types in each DRAGON cluster (Fig. 3b). (e) Overrepresentation analyses on gene signatures characterizing the Cre Control (6) and Lag3∆TM dominated clusters (3 + 4) was performed using KEGG pathways and the top 10 overrepresented in each genotype are shown. Enrichment ratio and –log10FDR (false discovery rate) are portrayed. (f) Heatmap of gene expression levels in the over-represented KEGG pathways.
Extended Data Fig. 6 Pseudotemporal analysis recapitulates the development of exhaustion in intra-islet CD8+ T cells and reveals key differences between Lag3ΔTM and Cre Controls.
Diffusion maps were constructed and pseudotemporal ordering was inferred (Methods) using single-cell RNAseq data described in Ext. Data Fig. 5. (a-d) CD8+ T cells from the islets and ndLN were isolated from 4 Cre Control and 4 Lag3∆TM 8-week NOD female mice and were subjected to 5’ paired single cell RNAseq (scRNAseq) and single cell T cell receptor sequencing (scTCRseq). Unless otherwise noted, red is representative of Lag3∆TM dominated clusters (3 + 4) and blue is representative of Cre Control dominated clusters (6). Diffusion component 1 and 2 portray the trajectory of CD8+ T cell differentiation. (a) Diffusion pseudotime colored by DRAGON cluster (Fig. 3b). (b-d) Differential gene expression as a function of diffusion pseudotime. Genes associated with early pseudotime (b), mid-pseudotime (c), and late pseudotime (d). Red corresponds to ORA markers of Lag3∆TM dominated clusters and blue is representative of Cre Control dominated cluster markers derived from ORA analysis. Two sided Pearson’s correlation was used to calculated the Pearson’s correlation coefficient where P < 2.2×10-16 (indicated as ****) in all cases.
Extended Data Fig. 7 TCR clonality in conjunction with diffusion pseudotime distinguish Lag3ΔTM and Cre Control samples.
(a-d) CD8 + T cells from the islets and ndLN were isolated from 4 Cre Control and 4 Lag3∆TM 8-week NOD female mice and were subjected to 5’ paired single cell RNAseq (scRNAseq) and single cell T cell receptor sequencing (scTCRseq). Red is representative of Lag3∆TM dominated clusters (3 + 4) and blue is representative of Cre Control dominated clusters (6). (a-b) 5’ scTCRseq was analyzed for frequency of expanded clones (a) and number of unique clones (b). Here, the line is the median, box is lower and upper quantiles (lower 25% and upper 25%), the upper whisker is the minimum of either the maximum value or the upper quartile plus 1.5 times the interquartile ranger. Bottom whisker is the maximum of the minimum or the first quartile minus 1.5 times in interquartile ranger. (c-d) Diffusion component 1 and 2 portray the trajectory of cellular development. (c) Diffusion pseudotime trajectory was divided into 5 clusters based on DC1 and DC2. Enrichment for islets begins in cluster 3, and clusters 4 and 5 constitute ~90% of cells derived from islets. (d) Expression of genes differentially regulated over time between Cre Control and Lag3∆TM in clusters enriched for cells derived from islet (that is clusters 3, 4 and 5).
Extended Data Fig. 8 Network analysis reveals differences in possible interactions between Lag3ΔTM and Cre Control.
(a-c) CD8+ T cells from the islets and ndLN were isolated from 4 Cre Control and 4 Lag3∆TM 8-week NOD female mice and were subjected to 5’ paired single cell RNAseq (scRNAseq) and single cell T cell receptor sequencing (scTCRseq). Unless otherwise noted, red is representative of Lag3∆TM seed genes (Clusters 3 + 4) and blue is representative of Cre Control seed genes (Cluster 6) (Supplementary Table 4). For subnetworks, all gene names are shown. (a) Protein subnetworks characterizing Cre Control (b) Protein subnetworks characterizing Lag3∆TM cells (c) Degree distribution for the different subnetworks showing that Cre Control have a higher frequency of networks with fewer connections.
Extended Data Fig. 9 LAG3 deletion has moderate impacts on proliferation, but phenotypically skews cells to an effector, rather than restrained phenotype.
The consequences of LAG3 deletion were evaluated by flow cytometry to phenotype intra-islet CD8 + T cells for survival, proliferation, and IR/exhaustion related marker expression. (a-e) Flow cytometry was performed on 8-week-old female Lag3∆TM and Cre Controls taking cells from ndLN, pLN and islets. Data points derived from islets having <40 tetramer+ cells were excluded. Tetramer staining in lymph nodes was negligible and never exceeded 40 Tetramer+ cells. (a) BrdU was injected intraperitoneally 12 hours prior to harvest, and percent BrdU, Ki67, cleaved Caspase 3 (p = .0496), and BCL2 were assessed by flow cytometry (2 independent experiments, n = 6-8 per genotype). (b) CD8 + T cells were labeled with cell trace violet, sorted into 96 well round bottom plate containing 0.05 ug/mL αCD3/CD28, and 200 U/mL IL-2 in cRPMI, and cultured for 60 hours and analyzed by flow cytometry (2 independent experiments, n = 6 per genotype). (c) IRs/markers of restraint (TIGIT, TCF1, PD1, and TOX) expression were quantified on total and tetramer positive CD8 + T cells (3 independent experiments, n = 13-15 per genotype Tetramer: TIGIT, PD1, TOX, p = .0473, 0.0473, 0.0096). (d) Percent expression of effector molecules CD44 (3 independent experiments, n = 13-15 per genotype ndLN, pLN, Islets p = .0016, 0.0007 .0037) and KLRG1 (1 independent experiment, n = 4-5 per genotype p = .036). (e) Percent expression of PD1 and TOX (Nrpv7 + p = .0259, InsB+ p = .0204), CD39 and Eomes (p = .03), double positive populations, markers of exhaustion, were monitored on bulk and tetramer positive ndLN, pLN, and intra-islet CD8 + T cells (3 independent experiments, n = 13-15 per genotype and 2 independent experiments, n = 5-6 per genotype, respectively). (a-e) Each data point corresponds to a single mouse. A two-sided nonparametric Mann-Whitney statistical test was preformed where P = * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. Unlabeled indicates not statistically significant. Graphs portray the median.
Extended Data Fig. 10 LAG3 deletion doesn’t affect single cytokine production or metabolic capacity.
The consequences of LAG3 deletion were evaluated by flow cytometry to phenotype intra-islet CD8+ T cells for cytokine production, metabolic capacity, and antigen specificity. (a-d) Flow cytometry was performed on 8-week-old female Lag3∆TM and Cre Controls taking cells from ndLN, pLN and islets. (a-b) lymphocytes were stimulated ex vivo for 5 hours with PMA, ionomycin, and brefeldin A and then assessed for cytokine production and degranulation. CD107a, GzmB, Tnfα and IFNγ were quantified (2 independent experiments n = 6-7 per genotype). Cytokine production is unchanged between genotypes, though dual cytokine production, an indicator of polyfunctionality, IFNγ+Gzmb+, is increased in Lag3∆TM (ndLN, pLN, Islet p = .0083, 0.035, 0.44) (b). (c) Lymphocytes were isolated from islets, ndLN, and pLN, cultured in serum free media for 37 degrees C in the presence of GlucoseCy5, CellROX, or MitoSOX, for 30 mins, surface stained including TMRE and MitoTracker, and analyzed by flow cytometry (2 independent experiments, n = 2-6 per genotype). Lag3L/L-YFP.NOD controls were included in this experiment to control for fluorescent protein expression that may overlap with metabolic markers. (a-c) Each data point corresponds to a single mouse. A two-sided nonparametric Mann-Whitney statistical test was preformed where P = * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. Unlabeled indicates not statistically significant. Graphs portray the median. (d) representative flow plots of tetramer staining in 8-week-old female Lag3∆TM and Cre Controls intra-islet CD8+ T cells.
Supplementary information
Source data
Source Data Fig. 1
Raw flow data.
Source Data Fig. 2
Raw flow data.
Source Data Fig. 3
Diabetes incidence table for Fig. 3a.
Source Data Fig. 4
Insulitis scoring and raw flow data.
Source Data Extended Data Fig. 1
Raw flow data.
Source Data Extended Data Fig. 3
Raw flow data.
Source Data Extended Data Fig. 4
Raw flow data.
Source Data Extended Data Fig. 5
Raw quantitative PCR data.
Source Data Extended Data Fig. 9
Raw flow data.
Source Data Extended Data Fig. 10
Raw flow data.
Rights and permissions
About this article
Cite this article
Grebinoski, S., Zhang, Q., Cillo, A.R. et al. Autoreactive CD8+ T cells are restrained by an exhaustion-like program that is maintained by LAG3. Nat Immunol 23, 868–877 (2022). https://doi.org/10.1038/s41590-022-01210-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-022-01210-5
This article is cited by
-
New insights into the stemness of adoptively transferred T cells by γc family cytokines
Cell Communication and Signaling (2023)
-
Novel strategies for cancer immunotherapy: counter-immunoediting therapy
Journal of Hematology & Oncology (2023)
-
The expression profiles of signature genes from CD103+LAG3+ tumour-infiltrating lymphocyte subsets predict breast cancer survival
BMC Medicine (2023)
-
LAG-3 as the third checkpoint inhibitor
Nature Immunology (2023)
-
From bench to bedside: targeting lymphocyte activation gene 3 as a therapeutic strategy for autoimmune diseases
Inflammation Research (2023)