Abstract
Mammalian circadian oscillators are built on a feedback loop in which the activity of the transcription factor CLOCK–BMAL1 is repressed by the PER–CRY complex. Here, we show that murine Per−/− fibroblasts display aberrant nucleosome occupancy around transcription start sites (TSSs) and at promoter-proximal and distal CTCF sites due to impaired histone H2A.Z deposition. Knocking out H2A.Z mimicked the Per null chromatin state and disrupted cellular rhythms. We found that endogenous mPER2 complexes retained CTCF as well as the specific H2A.Z-deposition chaperone YL1—a component of the ATP-dependent remodeler SRCAP and p400–TIP60 complex. While depleting YL1 or mutating chaperone-binding sites on H2A.Z lengthened the circadian period, H2A.Z deletion abrogated BMAL1 chromatin recruitment and promoted its proteasomal degradation. We propose that a PER2-mediated H2A.Z deposition pathway (1) compacts CLOCK–BMAL1 binding sites to establish negative feedback, (2) organizes circadian chromatin landscapes using CTCF and (3) bookmarks genomic loci for BMAL1 binding to impinge on the positive arm of the subsequent cycle.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Data sets generated by the lab are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE153939.
H2A.Z, H3K27me3 and H3K9me2 positions were obtained from GSE51505. H2A.Z coverage over several ZT points was obtained from GSE47145. PER1, PER2 and BMAL1 positions were obtained from GSE39860. CTCF sites were taken from GSE102997.
The reference mouse genome used in the study was mm10, gencode vM22 from UCSC (https://hgdownload.soe.ucsc.edu/goldenPath/mm10/bigZips/). Source data are provided with this paper.
References
Takahashi, J. S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017).
Duong, H. A., Robles, M. S., Knutti, D. & Weitz, C. J. A molecular mechanism for circadian clock negative feedback. Science 332, 1436–1439 (2011).
Kume, K. et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 (1999).
Padmanabhan, K., Robles, M. S., Westerling, T. & Weitz, C. J. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337, 599–602 (2012).
Sangoram, A. M. et al. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK–BMAL1-induced transcription. Neuron 21, 1101–13 (1998).
DeBruyne, J. P., Weaver, D. R. & Reppert, S. M. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci. 10, 543–545 (2007).
Siepka, S. M. et al. Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–23 (2007).
Godinho, S. I. et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900 (2007).
Busino, L. et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–4 (2007).
Lazar, M. A. in A Time for Metabolism and Hormones (eds. Sassone-Corsi, P. & Christen, Y.) 63–70 (Cham, 2016).
Albrecht, U. & Eichele, G. The mammalian circadian clock. Curr. Opin. Genet Dev. 13, 271–277 (2003).
Koike, N. et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012).
Yeung, J. et al. Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Genome Res. 28, 182–191 (2018).
Vollmers, C. et al. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 16, 833–45. (2012).
Lande-Diner, L., Boyault, C., Kim, J. Y. & Weitz, C. J. A positive feedback loop links circadian clock factor CLOCK–BMAL1 to the basic transcriptional machinery. Proc. Natl Acad. Sci. USA 110, 16021–6 (2013).
Duong, H. A. & Weitz, C. J. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat. Struct. Mol. Biol. 21, 126–32 (2014).
Brown, S. A. et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308, 693–6 (2005).
Ripperger, J. A. & Schibler, U. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–74 (2006).
Liu, X. et al. DNA replication is required for circadian clock function by regulating rhythmic nucleosome composition. Mol. Cell 67, 203–213 (2017).
Menet, J. S., Pescatore, S. & Rosbash, M. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev. 28, 8–13 (2014).
Mermet, J. et al. Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev. 32, 347–358 (2018).
Kim, Y. H. et al. Rev-erbα dynamically modulates chromatin looping to control circadian gene transcription. Science 359, 1274–1277 (2018).
Aguilar-Arnal, L. et al. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat. Struct. Mol. Biol. 20, 1206–13 (2013).
Zhao, H. et al. PARP1- and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol. Cell 59, 984–997 (2015).
xu, y. et al. long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression. PLoS Genet. 12, e1005992 (2016).
Papazyan, R., Zhang, Y. & Lazar, M. A. Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat. Struct. Mol. Biol. 23, 1045–1052 (2016).
Cauchy, P., Koch, F. & Andrau, J. C. Two possible modes of pioneering associated with combinations of H2A.Z and p300/CBP at nucleosome-occupied enhancers. Transcription 8, 179–184 (2017).
Nekrasov, M. et al. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat. Struct. Mol. Biol. 19, 1076–1083 (2012).
Soboleva, T. A., Nekrasov, M., Ryan, D. P. & Tremethick, D. J. Histone variants at the transcription start-site. Trends Genet. 30, 199–209 (2014).
Mylonas, C., Lee, C., Auld, A. L. & Cisse, L. A. Boyer II A dual role for H2A.Z.1 in modulating the dynamics of RNA polymerase II initiation and elongation. Nat. Struct. Mol. Biol. 28, 435–442 (2021).
Cole, L. et al. Multiple roles of H2A.Z in regulating promoter chromatin architecture in human cells. Nat. Commun. 12, 2524 (2021).
Hardy, S. & Robert, F. Random deposition of histone variants: a cellular mistake or a novel regulatory mechanism? Epigenetics 5, 368–372 (2010).
Fu, Y., Sinha, M., Peterson, C. L. & Weng, Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 4, e1000138 (2008).
Latrick, C. M. et al. Molecular basis and specificity of H2A.Z-H2B recognition and deposition by the histone chaperone YL1. Nat. Struct. Mol. Biol. 23, 309–316 (2016).
Liang, X. et al. Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1. Nat. Struct. Mol. Biol. 23, 317–323 (2016).
Punzeler, S. et al. Multivalent binding of PWWP2A to H2A.Z regulates mitosis and neural crest differentiation. EMBO J. 36, 2263–2279 (2017).
Gaucher, J., Montellier, E. & Sassone-Corsi, P. Molecular cogs: interplay between circadian clock and cell cycle. Trends Cell Biol. 28, 368–379 (2018).
Chao, H. W. et al. Circadian clock regulates hepatic polyploidy by modulating Mkp1-Erk1/2 signaling pathway. Nat. Commun. 8, 2238 (2017).
Albrecht, U., Bordon, A., Schmutz, I. & Ripperger, J. The multiple facets of Per2. Cold Spring Harb. Symp. Quant. Biol. 72, 95–104 (2007).
Dallmann, R. & Weaver, D. R. Altered body mass regulation in male mPeriod mutant mice on high-fat diet. Chronobiol. Int. 27, 1317–1328 (2010).
Allier, C. et al. Quantitative phase imaging of adherent mammalian cells: a comparative study. Biomed. Opt. Express 10, 2768–2783 (2019).
Allier, C. et al. Imaging of dense cell cultures by multiwavelength lens-free video microscopy. Cytom. Part A J. Int. Soc. Anal. Cytol. 91, 433–442 (2017).
Allier, C. et al. Lens-free video microscopy for the dynamic and quantitative analysis of adherent cell culture. Journal of visualized experiments. J. Vis. Exp. 56580 (2018).
Albrecht, U. et al. mPer1 and mPer2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms 16, 100–104 (2001).
Buschbeck, M. & Hake, S. B. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 18, 299–314 (2017).
Hockings, C. et al. Illuminating chromatin compaction in live cells and fixed tissues using SiR-DNA fluorescence lifetime. Preprint at bioRxiv https://doi.org/10.1101/2020.05.02.073536 (2020).
Jin, C. & Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21, 1519–1529 (2007).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Mieczkowski, J. et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 7, 11485 (2016).
Chaix, A., Zarrinpar, A. & Panda, S. The circadian coordination of cell biology. J. Cell Biol. 215, 15–25 (2016).
Cheema, M. S. & Ausio, J. The structural determinants behind the epigenetic role of histone variants. Genes 6, 685–713 (2015).
Kornmann, B., Schaad, O., Reinke, H., Saini, C. & Schibler, U. Regulation of circadian gene expression in liver by systemic signals and hepatocyte oscillators. Cold Spring Harb. Symp. Quant. Biol. 72, 319–30 (2007).
Lamia, K. A., Storch, K. F. & Weitz, C. J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl Acad. Sci. USA 105, 15172–7 (2008).
Wang, B., Kettenbach, A. N., Gerber, S. A., Loros, J. J. & Dunlap, J. C. Neurospora WC-1 recruits SWI/SNF to remodel frequency and initiate a circadian cycle. PLoS Genet. 10, e1004599 (2014).
Obri, A. et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505, 648–53 (2014).
Choi, J., Heo, K. & An, W. Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic Acids Res. 37, 5993–6007 (2009).
Kim, J. Y., Kwak, P. B. & Weitz, C. J. Specificity in circadian clock feedback from targeted reconstitution of the NuRD corepressor. Mol. Cell 56, 738–48 (2014).
Ju, D. et al. Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals. Sci. Transl. Med. 12, eaba0769 (2020).
Maier, B. et al. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 23, 708–718 (2009).
Subramanian, V. et al. H2A.Z: a molecular rheostat for transcriptional control. F1000Prime Rep. 7, 01 (2015).
Belotti, E. et al. H2A.Z is dispensable for both basal and activated transcription in post-mitotic mouse muscles. Nucleic Acids Res. 48, 4601–4613 (2020).
Hardy, S. et al. The euchromatic and heterochromatic landscapes are shaped by antagonizing effects of transcription on H2A.Z deposition. PLoS Genet. 5, e1000687 (2009).
Giaimo, B. D., Ferrante, F., Herchenrother, A., Hake, S. B. & Borggrefe, T. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 12, 37 (2019).
Holwerda, S. J. & de Laat, W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120369 (2013).
Braccioli, L. & de Wit, E. CTCF: a Swiss-army knife for genome organization and transcription regulation. Essays Biochem. 63, 157–165 (2019).
Wiechens, N. et al. The chromatin remodelling enzymes SNF2H and SNF2L position nucleosomes adjacent to CTCF and other transcription factors. PLoS Genet. 12, e1005940 (2016).
Trott, A. J. & Menet, J. S. Regulation of circadian clock transcriptional output by CLOCK:BMAL1. PLoS Genet. 14, e1007156 (2018).
Valdes-Mora, F. et al. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 22, 307–321 (2012).
Domaschenz, R., Kurscheid, S., Nekrasov, M., Han, S. & Tremethick, D. J. The histone variant H2A.Z is a master regulator of the epithelial–mesenchymal transition. Cell Rep. 21, 943–952 (2017).
Yang, B. et al. H2A.Z regulates tumorigenesis, metastasis and sensitivity to cisplatin in intrahepatic cholangiocarcinoma. Int. J. Oncol. 52, 1235–1245 (2018).
Ramanathan, C. et al. mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet. 14, e1007369 (2018).
Schmutz, I. et al. Protein phosphatase 1 (PP1) is a post-translational regulator of the mammalian circadian clock. PLoS ONE 6, e21325 (2011).
Littleton, E. S., Childress, M. L., Gosting, M. L., Jackson, A. N. & Kojima, S. Genome-wide correlation analysis to identify amplitude regulators of circadian transcriptome output. Sci. Rep. 10, 21839 (2020).
Kwon, I. et al. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol. Cell Biol. 26, 7318–7330 (2006).
Stratmann, M., Suter, D. M., Molina, N., Naef, F. & Schibler, U. Circadian Dbp transcription relies on highly dynamic BMAL1-CLOCK interaction with E boxes and requires the proteasome. Mol. Cell 48, 277–287 (2012).
Tamayo, A. G., Duong, H. A., Robles, M. S., Mann, M. & Weitz, C. J. Histone monoubiquitination by Clock–Bmal1 complex marks Per1 and Per2 genes for circadian feedback. Nat. Struct. Mol. Biol. 22, 759–766 (2015).
Segala, G., Bennesch, M. A., Pandey, D. P., Hulo, N. & Picard, D. Monoubiquitination of histone H2B blocks eviction of histone variant H2A.Z from inducible enhancers. Mol. Cell 64, 334–346 (2016).
Colino-Sanguino, Y. et al. A read/write mechanism connects p300 bromodomain function to H2A.Z acetylation. iScience 21, 773–788 (2019).
Zhao, X. et al. Nuclear receptors rock around the clock. EMBO Rep. 15, 518–528 (2014).
Kim, Y. H. et al. Rev-erbalpha dynamically modulates chromatin looping to control circadian gene transcription. Science 359, 1274–1277 (2018).
Mermet, J., Yeung, J. & Naef, F. Oscillating and stable genome topologies underlie hepatic physiological rhythms during the circadian cycle. PLoS Genet. 17, e1009350 (2021).
Wen, Z., Zhang, L., Ruan, H. & Li, G. Histone variant H2A.Z regulates nucleosome unwrapping and CTCF binding in mouse ES cells. Nucleic Acids Res. 48, 5939–5952 (2020).
Asher, G. et al. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142, 943–953 (2010).
Kettner, N. M. et al. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 30, 909–924 (2016).
Lee, C. C. Tumor suppression by the mammalian Period genes. Cancer Causes Control 17, 525–530 (2006).
Berta, D. G. et al. Deficient H2A.Z deposition is associated with genesis of uterine leiomyoma. Nature 596, 398–403 (2021).
Xu, Y. et al. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48, 723–733 (2012).
Huang, X., & Darzynkiewicz, Z. Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol. Biol. 314, 73–80 (2006).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Chen, K. et al. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 23, 341–351 (2013).
Kraushaar, D. C. et al. Erratum to: Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 17, 21 (2016).
Matthews, B. J. & Waxman, D. J. Computational prediction of CTCF/cohesin-based intra-TAD loops that insulate chromatin contacts and gene expression in mouse liver. eLife 7, e34077 (2018).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–5 (2010).
Anders, S., Reyes, A. & Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 (2012).
Herrmann, C., Avgousti, D. C. & Weitzman, M. D. Differential salt fractionation of nuclei to analyze chromatin-associated proteins from cultured mammalian cells. Bio. Protoc. 7, e2175 (2017).
Acknowledgements
The work was supported by an ATIP Avenir installation grant, an ENS emergence award, Projét Fondation ARC (PJA 20191209722) and a La Ligue contre le Cancer, Comité de l’Isère mono-équipe grant (R15026CC) to K. P. E. G. F. was funded by the ATIP Avenir postdoctoral fellowship, F. A. was supported by a grant from CLARA Oncostarter (CVPPRCAN000174) and K. T. was funded by an INSERM Plan Cancer in Physics (CP17067-00) grant to C. A. and K. P.. We would like to thank S. Yamazaki for providing us with PER-TKO cells, to D. Weaver for reaching out to S. Yamazaki; S. Dimitrov for access to the H2A.Z conditional mouse model; M.-P. Felder Schmittbuhl and U. Albrecht for Per1−/−Per2 Brdm−/− mice; S. Brown for Bmal1–luciferase lentiviral plasmid; A. Kramer for U2OS Bmal1–luc cells; C. Weitz for BMAL1 antibody; S. Romand for generating Per constructs; A. Ors for primer design and comments; M. Fackeure, S. Arenales and O. Bartle for technical help; B. Gillet and S. Hughes at the IGFL sequencing platform (PSI) for Illumina sequencing; and team members, A. Garces, Y. Ghavi-Helm and F. Leulier for discussions pertaining to the manuscript. We gratefully acknowledge support from C. Kabir at IGFL and the Pôle Scientifique de Modélisation Numérique at ENS de Lyon for computing resources. We particularly would like to extend our sincere gratitude to K. Koronowski and J. Smith, members of the laboratory of P. Sassone-Corsi, a pioneer in the field, for their help with shipping liver specimens from the Liver specific Bmal1-/- KO mice.
Author information
Authors and Affiliations
Contributions
K. P. conceived and directed the project with inputs from K. T., F. A. and E. G. F. K. T., F. A., E. G. F., R. S., I. S., D. L. and K. P. conducted the experiments. K. T. and R. S. performed all the bioinformatics analyses. T. S. and P.-S.W. generated and isolated tissues from liver-specific Bmal1−/− mice. K. P. wrote the manuscript with input from C. A., K. T., F. A., E. G. F., D. L., S. T., P.-S.W. and S. A. B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural and Molecular Biology thanks Paul Wade, and Ueli Schibler for their contribution to the peer review of this work. Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of cell lines by Sir-DNA dye incorporation.
A) Left panel, wildtype, Per DKO, Per TKO cells and mPer2 rescue fibroblasts were treated with DMSO or 1mM of siR-DNA live cell Fluorogenic Labelling Probe (SpirochromeSiR-DNA) and analyzed by flow cytometry (representative of N=3 experiments). Right panel, Results of three independent experiments are represented as mean±SD of the geometric mean. A one-tailed t-test was performed. B) Total genomic DNA from Per TKO cells, mPer2 rescue cell lines, wildtype fibroblasts and Per DKO cells was extracted and quantified. Results are expressed as mean±SD of N=3 independent experiments. A one-tailed t-test was performed. C) Sequential salt extraction (80-600mM NaCl) of wildtype and Per TKO nuclei were performed. Extracts as well as the final pellet were blotted for H2A.Z and H3. Results are representative of 4 experiments.
Extended Data Fig. 2 Nucleosome occupancy analysis (low MNase-seq) in H2A.Z DKO and Per TKO cells.
A) Tapestation profile of S0 nucleosomal fraction for wildtype, Per TKO and H2A.Z DKO cells prior to library preparation and sequencing. B) Average profile of low MNase-seq signal enrichment in wildtype Per TKO and H2AZ DKO cells at PER1 binding site, and at some repetitive elements (LTR, SINE and simple repeats), at TSS of expressed genes (solid lines) or non-expressed genes (dashed lines) (RNAseq analysis, this study, Extended Data Fig. 6), and around H2A.Z nucleosome peaks in heterochromatin at H3K9me2 or H3K27me3 loci. H2A.Z, H3K27me3 and H3K9me2. N= 2 biological replicates were performed. C) Average profiles of signal enrichment for low and high MNase-seq (top and bottom, respectively) in wildtype fibroblasts and Per TKO cells for genes classified in GO term gene cluster ‘Estrogen pathway’.
Extended Data Fig. 3 Nucleosome occupancy analysis (high MNase-seq) in H2A.Z DKO and Per TKO cells.
Average profile of high MNase-seq signal enrichment in wildtype, Per TKO and H2AZ DKO cells at BMAL1, PER1 and PER2 binding site, at TSS of all genes, at H3K9me2 and H3K27me3 marked heterochromatin and at the border of repetitive elements (LINE, LTR, Simple repeats and SINE).
Extended Data Fig. 4 Interdependence of PER2 and H2A.Z loading in chromatin.
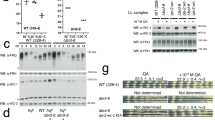
A) ChIP-seq profiles for H2A.Z, PER2 and BMAL1 in mouse liver at BMAL1-PER2 binding sites during the repressive phase. B) Western blot analysis showing the levels of H2A.Z in chromatin of different cell lines. H2A.Z DKO cells, two different cultures of Per TKO, mPer2-rescue or Bmal1−/− cell lines are represented. H3 was used as loading control. Results are representative of 3 experiments. C) Nuclear fractionation of cells into nucleoplasm (Np), chromatin fraction using MNase digestion (Ch1), chromatin fraction using MNase digestion with sonication (Ch2) and insoluble pellets post centrifugation (P1, P2). Fractions were analyzed for PER2, BMAL1, H2A.Z and H3. Results are representative of n>4 experiments. D) Bioluminescence profiles from U2OS cells stably expressing different shRNA constructs targeting YL1 (shYL1a (green), shYL1b (red) and shYL1c (purple)) compared to a scrambled control (shSCR). E) Images of cells at the end of lumicycle analysis (5X and 10X mag), Scale bar is 100 µm. Results are representative of 3 experiments. F) Immunoblot for H2A.Z in cells from the lumicycle at the end of the bioluminescence analysis, βActin was used as a loading control. Results are representative of 2 experiments. G) Western blot analysis for total cell extracts or for chromatin fraction for PER2, BMAL1 and H2A.Z on wildtype cells or following 3, 5 or 7 days of tamoxifen treatment. βActin or Histone H3 were used as loading controls. Results are representative of 4 experiments. H) Western blot analysis for total cell extracts or for the chromatin fraction for BMAL1, H2A.Z and H3 on wildtype cells (WT) and H2A.Z DKO treated with doxycycline to induce H2A.Z1. Non doxycycline treated cells were used as control. Results are representative of 2 experiments.
Extended Data Fig. 5 H2A.Z regulation of circadian period and amplitude.
A) Period determination in wildtype and H2A.Z DKO fibroblasts N=6 independent experiments (for for example, Fig. 5a), mean±SEM and p-values (t-test, paired, two-tailed) are indicated. B) Upper panel. Tamoxifen treated H2A.Z1 KO cells or H2A.Z2 KO cells (day 4+TAM) total RNA were subjected to RT-qPCR analysis for the expression of H2afz, H2afz, Bmal1 and Per2. The expression levels are normalized with Rps9 gene and compared to the wildtype cells. Results are expressed as fold change mean±SD in N=3 independent experiments. A two tailed paired t-test was performed (p-values<0.05 are indicated). Lower panel. Untreated wildtype cells (black traces), H2A.Z1 KO or H2A.Z2 KO cells (day 4+TAM, red traces) expressing a Bmal1:luciferase reporter were synchronized and bioluminescence was recorded for 5 days. C) Left panel. Bioluminescence recordings from wildtype fibroblasts, or rescue lines (following doxycycline mediated induction of H2A.Z1 expression in Tamoxifen treated cells). Right panel. Immunoblots for H2A.Z and H3 (loading control) from wildtype or H2A.Z DKO cells expressing H2A.Z1 rescue constructs as indicated above the blot. Results are representative of N=3 experiments. D) Phase contrast images of cells prior to and after real-time luminescence recordings for wildtype, H2A.Z DKO, and H2A.Z DKO rescued with H2A.Z1-Nter K5R mutant. Results are representative of N=3 experiments.
Extended Data Fig. 6 Transcriptomic analysis of H2A.Z DKO and Per TKO cells.
A) Illumina total RNASeq Pearson correlation plots of gene read counts for quadruplicate experiments for wildtype, H2A.Z DKO (+tamoxifen, day 4) and Per TKO cells. B) Volcano plot summarizing RNA-Seq data for Per TKO vs wildtype expression. DESeq analysis identified 5275 differentially expressed genes (>2 fold difference in expression; p-value <0.01 (DE test)). C) Volcano plot showing deregulated genes in Per TKO cells and focused on genes belonging to the GO term: P400-TIP60 SRCAP complex. In red, genes with >1.5 fold difference and p-value <0.01 (DE test). D) Log2 fold change and p-values (DE test) for some specific core clock genes from RNAseq (wildtype vs H2A.Z DKO) data are highlighted. E) Log fold change in expression between wildtype and H2A.Z DKO RNA for GO terms ‘ATP generation pathway’ (light grey) and ‘oxidoreductase activity on NAD(P)H’ (dark grey) pathways. Center line denotes the median value (50th percentile), while the box contains the 25th and 75th percentiles of the dataset. Mean is indicated by the cross. The whiskers mark the 5th and the 95th percentiles and values beyond these bounds are consider outliers in circles F) Volcano plot showing deregulated genes in H2A.Z DKO fibroblasts, for subset belonging to the GO term ‘regulation of circadian rhythms’. In red are highlighted genes with >2 fold difference and p-value <0.01(DE test).
Supplementary information
Supplementary Table 1
Number of sequences and statistics for the different sequencing datasets
Supplementary Table 2
ChIP–qPCR primers list
Supplementary Table 3
RT–qPCR primers list
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 1
Unprocessed Western Blots
Source Data Fig. 2
Unprocessed Western Blots
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 3
Unprocessed Western Blots
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 4
Unprocessed Western Blots
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 5
Unprocessed Western Blots
Source Data Fig. 6
Unprocessed Western Blots
Source Data Fig. 7
Statistical Source Data
Source Data Fig. 8
Statistical Source Data
Source Data Fig. 8
Unprocessed Western Blots
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 1
Unprocessed Western Blots
Source Data Extended Data Fig. 4
Unprocessed Western Blots
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 5
Unprocessed Western Blots
Source Data Extended Data Fig. 6
Statistical Source Data
Rights and permissions
About this article
Cite this article
Tartour, K., Andriani, F., Folco, E.G. et al. Mammalian PERIOD2 regulates H2A.Z incorporation in chromatin to orchestrate circadian negative feedback. Nat Struct Mol Biol 29, 549–562 (2022). https://doi.org/10.1038/s41594-022-00777-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00777-9
This article is cited by
-
Detecting abnormal cell behaviors from dry mass time series
Scientific Reports (2024)