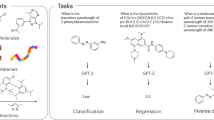
Abstract
Machine learning (ML) promises to tackle the grand challenges in chemistry and speed up the generation, improvement and/or ordering of research hypotheses. Despite the overarching applicability of ML workflows, one usually finds diverse evaluation study designs. The current heterogeneity in evaluation techniques and metrics leads to difficulty in (or the impossibility of) comparing and assessing the relevance of new algorithms. Ultimately, this may delay the digitalization of chemistry at scale and confuse method developers, experimentalists, reviewers and journal editors. In this Perspective, we critically discuss a set of method development and evaluation guidelines for different types of ML-based publications, emphasizing supervised learning. We provide a diverse collection of examples from various authors and disciplines in chemistry. While taking into account varying accessibility across research groups, our recommendations focus on reporting completeness and standardizing comparisons between tools. We aim to further contribute to improved ML transparency and credibility by suggesting a checklist of retro-/prospective tests and dissecting their importance. We envisage that the wide adoption and continuous update of best practices will encourage an informed use of ML on real-world problems related to the chemical sciences.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gawehn, E., Hiss, J. A., Brown, J. B. & Schneider, G. Advancing drug discovery via GPU-based deep learning. Expert Opin. Drug Discov. 13, 579–582 (2018).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 32, 8024–8035 (2019).
Abadi, M. et al. in Proc. 12th USENIX Conf. Operating Syst. Design Implement. 265–283 (USENIX Association, 2016).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Butler, K. T., Davies, D. W., Cartwright, H., Isayev, O. & Walsh, A. Machine learning for molecular and materials science. Nature 559, 547–555 (2018).
Sanchez-Lengeling, B. & Aspuru-Guzik, A. Inverse molecular design using machine learning: generative models for matter engineering. Science 361, 360–365 (2018).
Myszczynska, M. A. et al. Applications of machine learning to diagnosis and treatment of neurodegenerative diseases. Nat. Rev. Neurol. 16, 440–456 (2020).
Richens, J. G., Lee, C. M. & Johri, S. Improving the accuracy of medical diagnosis with causal machine learning. Nat. Commun. 11, 3923 (2020).
Yi, P. H., Malone, P., Lin, C. T. & Filice, R. W. Deep learning algorithms for interpretation of upper extremity radiographs: laterality and technologist initial labels as confounding factors. Am. J. Roentgenol. 218, 714–715 (2021).
Liu, X. et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digital Health 1, e271–e297 (2019).
Tschandl, P. et al. Human–computer collaboration for skin cancer recognition. Nat. Med. 26, 1229–1234 (2020).
de Almeida, A. F., Moreira, R. & Rodrigues, T. Synthetic organic chemistry driven by artificial intelligence. Nat. Rev. Chem. 3, 589–604 (2019).
Gromski, P. S., Henson, A. B., Granda, J. M. & Cronin, L. How to explore chemical space using algorithms and automation. Nat. Rev. Chem. 3, 119–128 (2019).
Schneider, P. et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 19, 353–364 (2020).
Strieth-Kalthoff, F., Sandfort, F., Segler, M. H. S. & Glorius, F. Machine learning the ropes: principles, applications and directions in synthetic chemistry. Chem. Soc. Rev. 49, 6154–6168 (2020).
Granda, J. M., Donina, L., Dragone, V., Long, D.-L. & Cronin, L. Controlling an organic synthesis robot with machine learning to search for new reactivity. Nature 559, 377–381 (2018).
Coley, C. W. et al. A robotic platform for flow synthesis of organic compounds informed by AI planning. Science 365, eaax1566 (2019).
Gómez-Bombarelli, R. et al. Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach. Nat. Mater. 15, 1120–1127 (2016).
Shamay, Y. et al. Quantitative self-assembly prediction yields targeted nanomedicines. Nat. Mater. 17, 361–368 (2018).
Reker, D., Hoyt, E. A., Bernardes, G. J. L. & Rodrigues, T. Adaptive optimization of chemical reactions with minimal experimental information. Cell Rep. Phys. Sci. 1, 100247 (2020).
Segler, M. H. S., Preuss, M. & Waller, M. P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555, 604–610 (2018).
Schreck, J. S., Coley, C. W. & Bishop, K. J. M. Learning retrosynthetic planning through simulated experience. ACS Cent. Sci. 5, 970–981 (2019).
Gómez-Bombarelli, R. et al. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 (2018).
Tu, K. H. et al. Machine learning predictions of block copolymer self-assembly. Adv. Mater. 32, 2005713 (2020).
Moret, M., Friedrich, L., Grisoni, F., Merk, D. & Schneider, G. Generative molecular design in low data regimes. Nat. Mach. Intell. 2, 171–180 (2020).
Yao, Z. et al. Inverse design of nanoporous crystalline reticular materials with deep generative models. Nat. Mach. Intell. 3, 76–86 (2021).
Segler, M. H. S., Kogej, T., Tyrchan, C. & Waller, M. P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 4, 120–131 (2018).
Gao, T. & Lu, W. Machine learning toward advanced energy storage devices and systems. iScience 24, 101936 (2021).
Severson, K. A. et al. Data-driven prediction of battery cycle life before capacity degradation. Nat. Energy 4, 383–391 (2019).
Rodrigues, T. et al. Machine intelligence decrypts β-lapachone as an allosteric 5-lipoxygenase inhibitor. Chem. Sci. 9, 6899–6903 (2018).
Conde, J. et al. Allosteric antagonist modulation of TRPV2 by piperlongumine impairs glioblastoma progression. ACS Cent. Sci. 7, 868–881 (2021).
Senior, A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020).
Wang, T. et al. Improved fragment sampling for ab initio protein structure prediction using deep neural networks. Nat. Mach. Intell. 1, 347–355 (2019).
Tian, Y. et al. Determining multi-component phase diagrams with desired characteristics using active learning. Adv. Sci. 8, 2003165 (2020).
Reker, D., Bernardes, G. J. L. & Rodrigues, T. Computational advances in combating colloidal aggregation in drug discovery. Nat. Chem. 11, 402–418 (2019).
Reker, D. et al. Computationally guided high-throughput design of self-assembling drug nanoparticles. Nat. Nanotech. 16, 725–733 (2021).
Timmreck, R. et al. Characterization of tandem organic solar cells. Nat. Photon. 9, 478–479 (2015).
Jones, D. T. Setting the standards for machine learning in biology. Nat. Rev. Mol. Cell Biol. 20, 659–660 (2019).
Walsh, I. et al. DOME: recommendations for supervised machine learning validation in biology. Nat. Mater. 18, 1122–1127 (2021).
Horstmeyer, R., Heintzmann, R., Popescu, G., Waller, L. & Yang, C. Standardizing the resolution claims for coherent microscopy. Nat. Photon. 10, 68–71 (2016).
Faria, M. et al. Minimum information reporting in bio–nano experimental literature. Nat. Nanotech. 13, 777–785 (2018).
Miernicki, M., Hofmann, T., Eisenberger, I., Kammer, F. V. D. & Praetorius, A. Legal and practical challenges in classifying nanomaterials according to regulatory definitions. Nat. Nanotech. 14, 208–216 (2019).
Aldrich, C. et al. The ecstasy and agony of assay interference compounds. ACS Cent. Sci. 3, 143–147 (2017).
Jain, A. N. & Nicholls, A. Recommendations for evaluation of computational methods. J. Computer Aided Mol. Des. 22, 133–139 (2008).
Artrith, N. et al. Best practices in machine learning for chemistry. Nat. Chem. 13, 505–508 (2021).
Alves, V. M. et al. SCAM detective: accurate predictor of small, colloidally aggregating molecules. J. Chem. Inf. Model. 60, 4056–4063 (2020).
Lee, K. et al. Combating small-molecule aggregation with machine learning. Cell Rep. Phys. Sci. 2, 100573 (2021).
Bender, A. & Cortés-Ciriano, I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 1: Ways to make an impact, and why we are not there yet. Drug Discov. Today 26, 511–524 (2021).
Bender, A. & Cortes-Ciriano, I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 2: A discussion of chemical and biological data. Drug Discov. Today 26, 1040–1052 (2021).
Brown, S. P., Muchmore, S. W. & Hajduk, P. J. Healthy skepticism: assessing realistic model performance. Drug Discov. Today 14, 420–427 (2009).
Robinson, M. C., Glen, R. C. & Lee, A. A. Validating the validation: reanalyzing a large-scale comparison of deep learning and machine learning models for bioactivity prediction. J. Computer Aided Mol. Des. 34, 717–730 (2020).
Cichońska, A. et al. Crowdsourced mapping of unexplored target space of kinase inhibitors. Nat. Commun. 12, 3307 (2021).
Brown, N., Fiscato, M., Segler, M. H. S. & Vaucher, A. C. GuacaMol: benchmarking models for de novo molecular design. J. Chem. Inf. Model. 59, 1096–1108 (2019).
Raji, I. D., Bender, E. M., Paullada, A., Denton, E. & Hanna, A. AI and the everything in the whole wide world benchmark. Preprint at arXiv https://arxiv.org/abs/2111.15366 (2021).
Renz, P., Rompaey, D. V., Wegner, J. K., Hochreiter, S. & Klambauer, G. On failure modes in molecule generation and optimization. Drug Discov. Today Technol. 32–33, 55–63 (2019).
Chen, L. et al. Hidden bias in the DUD-E dataset leads to misleading performance of deep learning in structure-based virtual screening. PLoS ONE 14, e0220113 (2019).
Wallach, I. & Heifets, A. Most ligand-based classification benchmarks reward memorization rather than generalization. J. Chem. Inf. Model. 58, 916–932 (2018).
Sieg, J., Flachsenberg, F. & Rarey, M. In need of bias control: evaluating chemical data for machine learning in structure-based virtual screening. J. Chem. Inf. Model. 59, 947–961 (2019).
Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Stanley, M. et al. in 35th Conf. Neural Inform. Process. Syst. Datasets Benchmarks Track (NeurIPS, 2021).
Thakkar, A., Kogej, T., Reymond, J.-L., Engkvist, O. & Bjerrum, E. J. Datasets and their influence on the development of computer assisted synthesis planning tools in the pharmaceutical domain. Chem. Sci. 11, 154–168 (2020).
Chen, G. et al. Alchemy: a quantum chemistry dataset for benchmarking AI models. Preprint at arXiv https://arxiv.org/abs/1906.09427 (2019).
Rodrigues, T. The good, the bad, and the ugly in chemical and biological data for machine learning. Drug Discov. Today Technol. 32–33, 3–8 (2019).
Heil, B. J. et al. Reproducibility standards for machine learning in the life sciences. Nat. Mater. 18, 1132–1135 (2021).
McCloskey, K. et al. Machine learning on DNA-encoded libraries: a new paradigm for hit finding. J. Med. Chem. 63, 8857–8866 (2020).
Giblin, K. A., Hughes, S. J., Boyd, H., Hansson, P. & Bender, A. Prospectively validated proteochemometric models for the prediction of small-molecule binding to bromodomain proteins. J. Chem. Inf. Model. 58, 1870–1888 (2018).
Mathai, N., Chen, Y. & Kirchmair, J. Validation strategies for target prediction methods. Brief. Bioinform. 21, 791–802 (2020).
Mitchell, J. B. O. Machine learning methods in chemoinformatics. Wiley Interdiscip. Rev. Comput. Mol. Sci. 4, 468–481 (2014).
Vishwakarma, G., Sonpal, A. & Hachmann, J. Metrics for benchmarking and uncertainty quantification: quality, applicability, and a path to best practices for machine learning in chemistry. Preprint at arXiv https://arxiv.org/abs/2010.00110 (2020).
Rosario, Z. D., Rupp, M., Kim, Y., Antono, E. & Ling, J. Assessing the frontier: active learning, model accuracy, and multi-objective candidate discovery and optimization. J. Chem. Phys. 153, 024112 (2020).
Schwaller, P., Gaudin, T., Lányi, D., Bekas, C. & Laino, T. “Found in translation”: predicting outcomes of complex organic chemistry reactions using neural sequence-to-sequence models. Chem. Sci. 9, 6091–6098 (2018).
Yu, T. & Zhu, H. Hyper-parameter optimization: a review of algorithms and applications. Preprint at arXiv https://arxiv.org/abs/2003.05689 (2020).
Schwaller, P., Vaucher, A. C., Laino, T. & Reymond, J.-L. Prediction of chemical reaction yields using deep learning. Mach. Learn. Sci. Technol. 2, 0115016 (2021).
Sandfort, F., Strieth-Kalthoff, F., Kühnemund, M., Beecks, C. & Glorius, F. A structure-based platform for predicting chemical reactivity. Chem 6, 1379–1390 (2020).
Scikit-learn Developers. Cross-validation: evaluating estimator performance. Scikit https://scikit-learn.org/stable/modules/cross_validation.html (2021).
Sheridan, R. P. Time-split cross-validation as a method for estimating the goodness of prospective prediction. J. Chem. Inf. Model. 53, 783–790 (2013).
Pesciullesi, G., Schwaller, P., Laino, T. & Reymond, J.-L. Transfer learning enables the molecular transformer to predict regio- and stereoselective reactions on carbohydrates. Nat. Commun. 11, 4874 (2020).
Ho, S. Y., Phua, K., Wong, L. & Goh, W. W. B. Extensions of the external validation for checking learned model interpretability and generalizability. Patterns 1, 100129 (2020).
Alexander, D. L. J., Tropsha, A. & Winkler, D. A. Beware of R2: simple, unambiguous assessment of the prediction accuracy of QSAR and QSPR models. J. Chem. Inf. Model. 55, 1316–1322 (2015).
Golbraikh, A. & Tropsha, A. Beware of q2! J. Mol. Graph. Model. 20, 269–276 (2002).
Consonni, V., Davide, B. & Todeschini, R. Comments on the definition of the Q2 parameter for QSAR validation. J. Chem. Inf. Model. 49, 1669–1678 (2009).
Derumigny, A. & Fermanian, J.-D. A classification point-of-view about conditional Kendall’s tau. Preprint at arXiv https://arxiv.org/abs/1806.09048 (2018).
Raeder, T., Forman, G. & Chawla, N. V. in Data Mining: Foundations and Intelligent Paradigms (eds Holmes, D. E. & Jain, L. C.) 315–331 (Springer, 2012).
Brown, J. B. Classifiers and their metrics quantified. Mol. Inf. 37, 1700127 (2018).
Beker, W., Wołos, A., Szymkuć, S. & Grzybowski, B. A. Minimal-uncertainty prediction of general drug-likeness based on Bayesian neural networks. Nat. Mach. Intell. 2, 457–465 (2020).
Perryman, A. L., Inoyama, D., Patel, J. S., Ekins, S. & Freundlich, J. S. Pruned machine learning models to predict aqueous solubility. ACS Omega 5, 16562–16567 (2020).
Schwaller, P. et al. Mapping the space of chemical reactions using attention-based neural networks. Nat. Mach. Intell. 3, 144–152 (2021).
Schwaller, P. et al. Predicting retrosynthetic pathways using transformer-based models and a hyper-graph exploration strategy. Chem. Sci. 11, 3316–3325 (2020).
Mo, Y. et al. Evaluating and clustering retrosynthesis pathways with learned strategy. Chem. Sci. 12, 1469–1478 (2021).
Talebian, S. et al. Facts and figures on materials science and nanotechnology progress and investment. ACS Nano 15, 15940–15952 (2021).
Olivecrona, M., Blaschke, T., Engkvist, O. & Chen, H. Molecular de-novo design through deep reinforcement learning. J. Cheminf. 9, 48 (2017).
Blaschke, T., Engkvist, O., Bajorath, J. & Chen, H. Memory-assisted reinforcement learning for diverse molecular de novo design. J. Cheminf. 12, 68 (2020).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Haghighi, S., Jasemi, M., Hessabi, S. & Zolanvari, A. PyCM: multiclass confusion matrix library in Python. J. Open Source Softw. 3, 729 (2018).
Beker, W., Gajewska, E. P., Badowski, T. & Grzybowski, B. A. Prediction of major regio-, site-, and diastereoisomers in Diels–Alder reactions by using machine-learning: the importance of physically meaningful descriptors. Angew. Chem. Int. Ed. 58, 4515–4519 (2019).
Has¨e, F., Roch, Lc. M., Kreisbeck, C. & Aspuru-Guzik, A. Phoenics: a Bayesian optimizer for chemistry. ACS Cent. Sci. 4, 1134–1145 (2018).
Nielsen, M. K., Ahneman, D. T., Riera, O. & Doyle, A. G. Deoxyfluorination with sulfonyl fluorides: navigating reaction space with machine learning. J. Am. Chem. Soc. 140, 5004–5008 (2018).
MacLeod, B. P. et al. Self-driving laboratory for accelerated discovery of thin-film materials. Sci. Adv. 6, eaaz8867 (2020).
Walters, W. P. & Murcko, M. Assessing the impact of generative AI on medicinal chemistry. Nat. Biotechnol. 38, 143–145 (2020).
Aickin, M. & Gensler, H. Adjusting for multiple testing when reporting research results: the Bonferroni vs Holm methods. Am. J. Public Health 86, 726–728 (1996).
Chuang, K. V. & Keiser, M. J. Adversarial controls for scientific machine learning. ACS Chem. Biol. 13, 2819–2831 (2018).
Ahneman, D. T., Estrada, J. G., Lin, S., Dreher, S. D. & Doyle, A. G. Predicting reaction performance in C–N cross-coupling using machine learning. Science 360, 186–190 (2018).
Chuang, K. V. & Keiser, M. J. Comment on “Predicting reaction performance in C–N cross-coupling using machine learning”. Science 362, eaat8603 (2018).
Schwaller, P. et al. Molecular transformer: a model for uncertainty-calibrated chemical reaction prediction. ACS Cent. Sci. 5, 1572–1583 (2019).
Maragakis, P., Nisonoff, H., Cole, B. & Shaw, D. E. A deep-learning view of chemical space designed to facilitate drug discovery. J. Chem. Inf. Model. 60, 4487–4496 (2020).
Zahrt, A. F. et al. Prediction of higher-selectivity catalysts by computer-driven workflow and machine learning. Science 363, eaau5631 (2019).
Reid, J. P., Proctor, R. S. J., Sigman, M. S. & Phipps, R. J. Predictive multivariate linear regression analysis guides successful catalytic enantioselective Minisci reactions of diazines. J. Am. Chem. Soc. 141, 19178–19185 (2019).
Brix, K. V., DeForest, D. K., Tear, L., Grose, M. & Adam, W. J. Use of multiple linear regression models for setting water quality criteria for copper: a complementary approach to the biotic ligand model. Environ. Sci. Technol. 51, 5182–5192 (2017).
Toste, F. D., Sigman, M. S. & Miller, S. J. Pursuit of noncovalent interactions for strategic site-selective catalysis. Acc. Chem. Res. 50, 609–615 (2017).
Reid, J. P. & Sigman, M. S. Holistic prediction of enantioselectivity in asymmetric catalysis. Nature 571, 343–348 (2019).
Zahrt, A. F., Athavale, S. V. & Denmark, S. E. Quantitative structure–selectivity relationships in enantioselective catalysis: past, present, and future. Chem. Rev. 120, 1620–1689 (2020).
Rodrigues, T. Deriving intuition in catalyst design with machine learning. Chem 8, 15–17 (2022).
Liu, B. et al. Retrosynthetic reaction prediction using neural sequence-to-sequence models. ACS Cent. Sci. 3, 1103–1113 (2017).
Dai, H., Li, C., Coley, C. W., Dai, B. & Song, L. Retrosynthesis prediction with conditional graph logic network. Preprint at arXiv https://arxiv.org/abs/2001.01408 (2020).
Vaucher, A. C. et al. Inferring experimental procedures from text-based representations of chemical reactions. Nat. Commun. 12, 2573 (2021).
Gillet, V. J., Willett, P. & Bradshaw, J. Identification of biological activity profiles using substructural analysis and genetic algorithms. J. Chem. Inf. Comput. Sci. 38, 165–179 (1998).
Edgar, S. J., Holliday, J. D. & Willett, P. Effectiveness of retrieval in similarity searches of chemical databases: a review of performance measures. J. Mol. Graph. Model. 18, 343–357 (2000).
Schneider, G. & Böhm, H.-J. Virtual screening and fast automated docking methods. Drug Discov. Today 7, 64–70 (2002).
Coley, C. W., Rogers, L., Green, W. H. & Jensen, K. F. Computer-assisted retrosynthesis based on molecular similarity. ACS Cent. Sci. 3, 1237–1245 (2017).
Capecchi, A., Probst, D. & Reymond, J.-L. One molecular fingerprint to rule them all: drugs, biomolecules, and the metabolome. J. Cheminf. 12, 43 (2020).
Rodrigues, T., Almeida, B. P. D., Barbosa-Morais, N. L. & Bernardes, G. J. L. Dissecting celastrol with machine learning to unveil dark pharmacology. Chem. Commun. 55, 6369–6372 (2019).
Rodrigues, T. et al. De novo fragment design for drug discovery and chemical biology. Angew. Chem. Int. Ed. 54, 15079–15083 (2015).
Häse, F., Roch, L. M., Friederich, P. & Aspuru-Guzik, A. Designing and understanding light-harvesting devices with machine learning. Nat. Commun. 11, 4587 (2020).
Zhavoronkov, A. et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 37, 1038–1040 (2019).
Moret, M., Helmstädter, M., Grisoni, F., Schneider, G. & Merk, D. Beam search for automated design and scoring of novel ROR ligands with machine intelligence. Angew. Chem. Int. Ed. 60, 19477–19482 (2021).
Kearnes, S. Pursuing a prospective perspective. Trends Chem. 3, 77–79 (2021).
Deringer, V. L. et al. Origins of structural and electronic transitions in disordered silicon. Nature 589, 59–64 (2021).
Porwol, L. et al. An autonomous chemical robot discovers the rules of inorganic coordination chemistry without prior knowledge. Angew. Chem. Int. Ed. 59, 11256–11261 (2020).
Kurczab, R., Smusz, S. & Bojarski, A. J. The influence of negative training set size on machine learning-based virtual screening. J. Cheminf. 6, 32 (2014).
Lewis, R. A., Ertl, P., Schneider, N. & Stiefl, N. Reducing the concepts of data science and machine learning to tools for the bench chemist. Chimia 73, 1001–1005 (2019).
Reutlinger, M., Rodrigues, T., Schneider, P. & Schneider, G. Multi-objective molecular de novo design by adaptive fragment prioritization. Angew. Chem. Int. Ed. 53, 4244–4248 (2014).
Anders, C. J., Montavon, G., Samek, W. & Müller, K.-R. in Explainable AI: Interpreting, Explaining and Visualizing Deep Learning (eds Samek, W., Montavon, G., Vedaldi, A., Hansen, L. K. & Müller, K.-R.) 297–309 (Springer, 2019).
Jiménez-Luna, J., Grisoni, F. & Schneider, G. Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2, 573–584 (2020).
Sheridan, R. P. Interpretation of QSAR models by coloring atoms according to changes in predicted activity: how robust is it? J. Chem. Inf. Model. 59, 1324–1337 (2019).
Matveieva, M. & Polishchuk, P. Benchmarks for interpretation of QSAR models. J. Cheminf. 13, 41 (2021).
Lundberg, S. M. et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2, 749–760 (2018).
Ribeiro, M. T., Singh, S. & Guestrin, C. “Why should I trust you?”: explaining the predictions of any classifier. Preprint at arXiv https://arxiv.org/abs/1602.04938 (2016).
Gao, H. et al. Using machine learning to predict suitable conditions for organic reactions. ACS Cent. Sci. 4, 1465–1476 (2018).
Zhong, M. et al. Accelerated discovery of CO2 electrocatalysts using active machine learning. Nature 581, 178–184 (2020).
Riniker, S. & Landrum, G. A. Similarity maps — a visualization strategy for molecular fingerprints and machine-learning methods. J. Cheminf. 5, 43 (2013).
Friederich, P., Krenn, M., Tamblyn, I. & Aspuru-Guzik, A. Scientific intuition inspired by machine learning generated hypotheses. Mach. Learn. Sci. Technol. 2, 025027 (2021).
Webel, H. E. et al. Revealing cytotoxic substructures in molecules using deep learning. J. Computer Aided Mol. Des. 34, 731–746 (2020).
Singh, S. et al. A unified machine-learning protocol for asymmetric catalysis as a proof of concept demonstration using asymmetric hydrogenation. Proc. Natl Acad. Sci. USA 117, 1339–1345 (2020).
Coley, C. W. et al. A graph-convolutional neural network model for the prediction of chemical reactivity. Chem. Sci. 10, 370–377 (2019).
Reker, D. & Schneider, G. Active-learning strategies in computer-assisted drug discovery. Drug Discov. Today 20, 458–465 (2015).
Reutlinger, M. et al. Chemically Advanced Template Search (CATS) for scaffold-hopping and prospective target prediction for ‘orphan’ molecules. Mol. Inf. 32, 133–138 (2013).
Reker, D., Schneider, P. & Schneider, G. Multi-objective active machine learning rapidly improves structure–activity models and reveals new protein–protein interaction inhibitors. Chem. Sci. 7, 3919–3927 (2016).
Schwaller, P., Hoover, B., Reymond, J.-L., Strobelt, H. & Laino, T. Extraction of organic chemistry grammar from unsupervised learning of chemical reactions. Sci. Adv. 7, eabe4166 (2021).
Burger, B. et al. A mobile robotic chemist. Nature 583, 237–241 (2020).
Gromski, P. S., Granda, J. M. & Cronin, L. Universal chemical synthesis and discovery with ‘The Chemputer’. Trends Chem. 2, 4–12 (2020).
Turing, A. M. Computing machinery and intelligence. Mind 56, 433–560 (1950).
Mikulak-Klucznik, B. et al. Computational planning of the synthesis of complex natural products. Nature 588, 83–88 (2020).
Duros, V. et al. Human versus robots in the discovery and crystallization of gigantic polyoxometalates. Angew. Chem. Int. Ed. 56, 10815–10820 (2017).
Klucznik, T. et al. Efficient syntheses of diverse, medicinally relevant targets planned by computer and executed in the laboratory. Chem 4, 522–532 (2018).
Shields, B. J. et al. Bayesian reaction optimization as a tool for chemical synthesis. Nature 590, 89–96 (2021).
Polykovskiy, D. et al. Molecular Sets (MOSES): a benchmarking platform for molecular generation models. Front. Pharmacol. 11, 1931 (2020).
Arús-Pous, J. et al. Exploring the GDB-13 chemical space using deep generative models. J. Cheminf. 11, 20 (2019).
Mysinger, M. M., Carchia, M., Irwin, J. J. & Shoichet, B. K. Directory of Useful Decoys, Enhanced (DUD-E): better ligands and decoys for better benchmarking. J. Med. Chem. 55, 6582–6594 (2012).
Lowe, D. M. Extraction of Chemical Structures and Reactions from the Literature. Thesis, Univ. Cambridge (2012).
Axelrod, S. & Gómez-Bombarelli, R. GEOM: energy-annotated molecular conformations for property prediction and molecular generation. Preprint at arXiv https://arxiv.org/abs/2006.05531 (2020).
Wang, R., Fang, X., Lu, Y., Yang, C.-Y. & Wang, S. The PDBbind database: methodologies and updates. J. Med. Chem. 48, 4111–4119 (2005).
García-Ortegón, M. et al. DOCKSTRING: easy molecular docking yields better benchmarks for ligand design. Preprint at arXiv https://arxiv.org/abs/2110.15486 (2021).
Sun, J. et al. ExCAPE-DB: an integrated large scale dataset facilitating big data analysis in chemogenomics. J. Cheminf. 9, 17 (2017).
Segler, M. H. S. & Waller, P. P. Neural-symbolic machine learning for retrosynthesis and reaction prediction. Chem. Eur. J. 23, 5966–5971 (2017).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at arXiv https://arxiv.org/abs/1802.03426 (2020).
Acknowledgements
T.R. acknowledges FCT Portugal for funding (CEECIND/00684/2018). T.R. thanks colleagues for discussions on the topic presented here over the years. D. Reker is acknowledged for providing access to original data discussed in the manuscript. T.R., O.E. and A.B. acknowledge that not all suggested evaluation studies might simultaneously be found in their own original research manuscripts. We thank M. Thomas and M. Garcia-Ortegon for help with Table 1.
Author information
Authors and Affiliations
Contributions
All authors contributed to the discussion and writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
T.R. is a co-founder and shareholder of TargTex and has acted as consultant to the pharmaceutical industry. A.B. is a co-founder and shareholder of Healx, PharmEnable and Terra Lumina and acts as a consultant to various pharmaceutical companies.
Peer review
Peer review information
Nature Reviews Chemistry thanks F. Grisoni and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
DOCKSTRING: https://github.com/dockstring/dockstring
DUD-E: http://dude.docking.org/
ExCAPE: https://solr.ideaconsult.net/search/excape/
FS-Mol: https://github.com/microsoft/FS-Mol
GDB-13: https://gdb.unibe.ch/downloads/
GEOM: https://github.com/learningmatter-mit/geom
GuacaMol: https://github.com/BenevolentAI/guacamol
Kaggle competitions: http://www.kaggle.com/
MoleculeNet: https://moleculenet.org/
MOSES: https://github.com/molecularsets/moses
PDBbind: http://www.pdbbind.org.cn/
RXNMapper: http://rxnmapper.ai/
SAMPL blind challenges: http://www.samplchallenges.org/
USPTO: https://figshare.com/articles/dataset/Chemical_reactions_from_US_patents_1976-Sep2016_/5104873
Supplementary information
Rights and permissions
About this article
Cite this article
Bender, A., Schneider, N., Segler, M. et al. Evaluation guidelines for machine learning tools in the chemical sciences. Nat Rev Chem 6, 428–442 (2022). https://doi.org/10.1038/s41570-022-00391-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41570-022-00391-9
This article is cited by
-
Predictive Minisci late stage functionalization with transfer learning
Nature Communications (2024)
-
Artificial molecular pumps
Nature Reviews Methods Primers (2024)
-
Tackling assay interference associated with small molecules
Nature Reviews Chemistry (2024)
-
On the difficulty of validating molecular generative models realistically: a case study on public and proprietary data
Journal of Cheminformatics (2023)
-
SIMPD: an algorithm for generating simulated time splits for validating machine learning approaches
Journal of Cheminformatics (2023)