Abstract
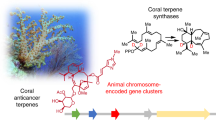
Octocorals are major contributors of terpenoid chemical diversity in the ocean. Natural products from other sessile marine animals are primarily biosynthesized by symbiotic microbes rather than by the host. Here, we challenge this long-standing paradigm by describing a monophyletic lineage of animal-encoded terpene cyclases (TCs) ubiquitous in octocorals. We characterized 15 TC enzymes from nine genera, several of which produce precursors of iconic coral-specific terpenoids, such as pseudopterosin, lophotoxin and eleutherobin. X-ray crystallography revealed that coral TCs share conserved active site residues and structural features with bacterial TCs. The identification of coral TCs enabled the targeted identification of the enzyme that constructs the coral-exclusive capnellane scaffold. Several TC genes are colocalized with genes that encode enzymes known to modify terpenes. This work presents an example of biosynthetic capacity in the kingdom Animalia that rivals the chemical complexity generated by plants, unlocking the biotechnological potential of octocorals for biomedical applications.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Shotgun sequencing reads for C. imbricata are available at NCBI SRA under project accession PRJNA761189. The crystal structure of ErTC-2 has been deposited at PDB under the accession number 7S5L. NMR and electron ionization MS spectra of characterized compounds are available in the Supplementary Information. Structure elucidation data are shown in Supplementary Note 1. Sequences used in this study are listed in Supplementary Note 2. Accession numbers of proteins used for HMM building are listed in Supplementary Table 1. Accession data for the literature genomic and transcriptomic data used in this study are summarized in Supplementary Tables 2 and 3, respectively. Accession data for raw genomic data used for mapping are listed in Supplementary Table 4. Accession numbers for proteins used for the phylogenetic analysis are listed in Supplementary Table 10. The PDB accession numbers for reference protein structures in Fig. 3 are 4LZ0, 3P5R and 5JA0.
References
Christianson, D. W. Structural and chemical biology of terpenoid cyclases. Chem. Rev. 117, 11570–11648 (2017).
Karunanithi, P. S. & Zerbe, P. Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front. Plant Sci. 10, 1166 (2019).
Dickschat, J. S. Bacterial terpene cyclases. Nat. Prod. Rep. 33, 87–110 (2016).
Quin, M. B., Flynn, C. M. & Schmidt-Dannert, C. Traversing the fungal terpenome. Nat. Prod. Rep. 31, 1449–1473 (2014).
Chen, X. et al. Terpene synthase genes in eukaryotes beyond plants and fungi: occurrence in social amoebae. Proc. Natl Acad. Sci. USA 113, 12132–12137 (2016).
Schiff, P. B., Fant, J. & Horwitz, S. B. Promotion of microtubule assembly in vitro by taxol. Nature 277, 665–667 (1979).
Eckstein-Ludwig, U. et al. Artemisinins target the SERCA of Plasmodium falciparum. Nature 424, 957–961 (2003).
Lyu, C. et al. CMNPD: a comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 49, D509–D515 (2021).
Giordano, G. et al. Volatile secondary metabolites as aposematic olfactory signals and defensive weapons in aquatic environments. Proc. Natl Acad. Sci. USA 114, 3451–3456 (2017).
Berrue, F. & Kerr, R. G. Diterpenes from gorgonian corals. Nat. Prod. Rep. 26, 681–710 (2009).
Li, G., Dickschat, J. S. & Guo, Y.-W. Diving into the world of marine 2,11-cyclized cembranoids: a summary of new compounds and their biological activities. Nat. Prod. Rep. 37, 1367–1383 (2020).
Look, S. A., Fenical, W., Jacobs, R. S. & Clardy, J. The pseudopterosins: anti-inflammatory and analgesic natural products from the sea whip Pseudopterogorgia elisabethae. Proc. Natl Acad. Sci. USA 83, 6238–6240 (1986).
Hassan, H. M. et al. Pachycladins A–E, prostate cancer invasion and migration inhibitory eunicellin-based diterpenoids from the red sea soft coral Cladiella pachyclados. J. Nat. Prod. 73, 848–853 (2010).
Fenical, W., Okuda, R. K., Bandurraga, M. M., Culver, P. & Jacobs, R. S. Lophotoxin: a novel neuromuscular toxin from Pacific sea whips of the genus Lophogorgia. Science 212, 1512–1514 (1981).
Wilson, M. C. et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014).
Smith, T. E. et al. Accessing chemical diversity from the uncultivated symbionts of small marine animals. Nat. Chem. Biol. 14, 179–185 (2018).
Sudek, S. et al. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 70, 67–74 (2007).
Torres, J. P. & Schmidt, E. W. The biosynthetic diversity of the animal world. J. Biol. Chem. 294, 17684–17692 (2019).
Shinzato, C. et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323 (2011).
McCauley, E. P. et al. Highlights of marine natural products having parallel scaffolds found from marine-derived bacteria, sponges, and tunicates. J. Antibiot. 73, 504–525 (2020).
Mydlarz, L. D., Jacobs, R. S., Boehnlein, J. & Kerr, R. G. Pseudopterosin biosynthesis in Symbiodinium sp., the dinoflagellate symbiont of Pseudopterogorgia elisabethae. Chem. Biol. 10, 1051–1056 (2003).
Frenz-Ross, J. L., Enticknap, J. J. & Kerr, R. G. The effect of bleaching on the terpene chemistry of Plexaurella fusifera: evidence that zooxanthellae are not responsible for sesquiterpene production. Mar. Biotechnol. 10, 572–578 (2008).
Kohl, A. C. & Kerr, R. G. Identification and characterization of the pseudopterosin diterpene cyclase, elisabethatriene synthase, from the marine gorgonian, Pseudopterogorgia elisabethae. Arch. Biochem. Biophys. 424, 97–104 (2004).
Wei, G. et al. Terpene biosynthesis in red algae is catalyzed by microbial type but not typical plant terpene synthases. Plant Physiol. 179, 382–390 (2019).
Yamada, Y. et al. Terpene synthases are widely distributed in bacteria. Proc. Natl Acad. Sci. USA 112, 857–862 (2015).
Mistry, J. et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021).
Jeon, Y. et al. The draft genome of an octocoral, Dendronephthya gigantea. Genome Biol. Evol. 11, 949–953 (2019).
Huber, T., Weisheit, L. & Magauer, T. Synthesis of Xenia diterpenoids and related metabolites isolated from marine organisms. Beilstein J. Org. Chem. 11, 2521–2539 (2015).
Hu, M., Zheng, X., Fan, C. -M. & Zheng, Y. Lineage dynamics of the endosymbiotic cell type in the soft coral Xenia. Nature 582, 534–538 (2020).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Li, R. et al. Reprogramming the chemodiversity of terpenoid cyclization by remolding the active site contour of epi-isozizaene synthase. Biochemistry 53, 1155–1168 (2014).
Köksal, M., Jin, Y., Coates, R. M., Croteau, R. & Christianson, D. W. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 469, 116–120 (2011).
Park, J., Zielinski, M., Magder, A., Tsantrizos, Y. S. & Berghuis, A. M. Human farnesyl pyrophosphate synthase is allosterically inhibited by its own product. Nat. Commun. 8, 14132 (2017).
Lancaster, J. et al. De novo formation of an aggregation pheromone precursor by an isoprenyl diphosphate synthase-related terpene synthase in the harlequin bug. Proc. Natl Acad. Sci. USA 115, E8634–E8641 (2018).
McFadden, C. S. et al. Phylogenomics, origin, and diversification of anthozoans (phylum Cnidaria). Syst. Biol. 70, 635–647 (2021).
Morris, J. L. et al. The timescale of early land plant evolution. Proc. Natl Acad. Sci. USA 115, E2274–E2283 (2018).
Medema, M. H., de Rond, T. & Moore, B. S. Mining genomes to illuminate the specialized chemistry of life. Nat. Rev. Genet. 22, 553–571 (2021).
Nützmann, H. -W., Huang, A. & Osbourn, A. Plant metabolic clusters—from genetics to genomics. New Phytol. 211, 771–789 (2016).
De La Peña, R. & Sattely, E. S. Rerouting plant terpene biosynthesis enables momilactone pathway elucidation. Nat. Chem. Biol. 17, 205–212 (2021).
Morris, L. A., Jaspars, M., Adamson, K., Woods, S. & Wallace, H. M. The capnellenes revisited: new structures and new biological activity. Tetrahedron 54, 12953–12958 (1998).
Jean, Y.-H. et al. Capnellene, a natural marine compound derived from soft coral, attenuates chronic constriction injury-induced neuropathic pain in rats. Br. J. Pharmacol. 158, 713–725 (2009).
Ayanoglu, E., Gebreyesus, T., Beechan, C. M., Djerassi, C. & Kaisin, M. Terpenoids LXXV. Δ9(12)-Capnellene, a new sesquiterpene hydrocarbon from the soft coral Capnella imbricata. Tetrahedron Lett. 19, 1671–1674 (1978).
DeLeo, D. M. et al. Gene expression profiling reveals deep-sea coral response to the Deepwater Horizon oil spill. Mol. Ecol. 27, 4066–4077 (2018).
Todaro, D. & Watson, G. M. Force-dependent discharge of nematocysts in the sea anemone Haliplanella luciae (Verrill). Biol. Open 1, 582–587 (2012).
Woodside, A. B., Huang, Z. & Poulter, C. D. Trisammonium geranyl diphosphate. Org. Synth. 66, 211 (1988).
Jo Davisson, V., Woodside, A. B. & Dale Poulter, C. D. Synthesis of allylic and homoallylic isoprenoid pyrophosphates. Methods Enzymol. 110, 130–144 (1985).
Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Priyam, A. et al. Sequenceserver: a modern graphical user interface for custom BLAST databases. Mol. Biol. Evol. 36, 2922–2924 (2019).
Lassmann, T., Frings, O. & Sonnhammer, E. L. L. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 37, 858–865 (2009).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Skubák, P. & Pannu, N. S. Automatic protein structure solution from weak X-ray data. Nat. Commun. 4, 2777 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
Cowtan, K. Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 (2010).
Skubák, P., Waterreus, W.-J. & Pannu, N. S. Multivariate phase combination improves automated crystallographic model building. Acta Crystallogr. D Biol. Crystallogr. 66, 783–788 (2010).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Biol. Crystallogr. 75, 861–877 (2019).
Pont-Kingdon, G. A. et al. A coral mitochondrial mutS gene. Nature 375, 109–111 (1995).
France, S. C. & Hoover, L. L. DNA sequences of the mitochondrial COI gene have low levels of divergence among deep-sea octocorals (Cnidaria: Anthozoa). Hydrobiologia 471, 149–155 (2002).
Sánchez, J. A., McFadden, C. S., France, S. C. & Lasker, H. R. Molecular phylogenetic analyses of shallow-water Caribbean octocorals. Mar. Biol. 142, 975–987 (2003).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 19, 455–477 (2012).
Miller, I. J. et al. Autometa: automated extraction of microbial genomes from individual shotgun metagenomes. Nucleic Acids Res. 47, e57 (2019).
Acknowledgements
We thank our University of California San Diego colleagues W. Fenical, L. Aluwihare and B. Duggan for critical feedback, access to GC–MS equipment and assistance with NMR measurements, respectively, C. Delbeek (California Academy of Sciences) for access to C. imbricata, J. Keasling and J. Blake-Hedges (University of California Berkeley) for assisting in genome sequencing, J. Bailey and the University of California San Diego Crystallography Facility for collecting the X-ray diffraction data used for phasing and J.P. Noel and G. Louie (Salk Institute for Biological Studies) for beamtime coordination. Use of the Stanford Synchrotron Radiation Lightsource is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-AC02-76SF00515, the Department of Energy Office of Biological and Environmental Research and the NIH and NIGMS (including P41GM103393). This work was supported by National Institutes of Health awards F32GM129960 to T.d.R. and R01GM085770 to B.S.M. and a Leopoldina postdoctoral fellowship (LPDS 2019-04) to I.B.
Author information
Authors and Affiliations
Contributions
I.B., T.d.R. and B.S.M. conceived the project, designed the experiments, analyzed the data and wrote the paper with input from all authors. I.B. performed the protein expression and purification, enzymatic assays, product analyses and data analyses with help from T.d.R. who additionally performed the C. imbricata experiments. P.Y.-T.C. performed crystallography experiments and subsequent data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Thu-Thuy Dang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Radial depiction of the phylogenetic tree from Fig. 2.
The IDS clade is used as outgroup and bootstrap values for the branches are shown. The representative enzymes used for structure comparison in Fig. 3 are indicated by arrows.
Extended Data Fig. 2 Examples of octocoral derived terpenoid structures sharing a common terpene carbon backbone8.
Structures marked with an asterisk have been found as both enantiomers.
Extended Data Fig. 3 Genetic context of octocoral TCs.
Mapped transcripts (top), gene annotations (middle) and mapped long-reads (bottom) for the indicated regions are shown. a) Context of XsTC-1 from Xenia sp. b) Context of DgTC-1 from D. gigantea. c) Context of DgTC-2 from D. gigantea. All genes are coded on long eukaryotic contigs (see Supplementary Table 4) with continuous long read coverage between 60 and 130 (Oxford Nanopore for a, PacBio for b and c). While the expression of DgTC-1 and DgTC-2 is low, XsTC-1 is highly expressed. For gene IDs see Supplementary Tables 5-7. The co-occurrence of two different bases in the same position (marked by colors) results from the heterozygosity of D. gigantea and Xenia sp.
Extended Data Fig. 4 Octocoral gene clusters.
Genomic contigs from Xenia sp., Renilla muelleri and Trachythela sp. show the physical co-localization of TCs with genes coding for CYP and SHD enzymes. Genes on the Xenia scaffold that are marked in grey are either annotated as unknown proteins or show similarity to enzymes with non-biosynthetic functions. CYP = cytochrome P450 monooxygenase, IDS = isoprenyl diphosphate synthase, SDH = short-chain dehydrogenase, TC = terpene cyclase.
Extended Data Fig. 5 Putative biochemical pathway from 7 to oxidized Xenia diterpenoids.
A: Putative terpenoid cluster including Cytochrome P450 genes and short chain dehydrogenase genes additionally to the characterized terpene cyclase XsTC-1, which produces 7. B: Putative overview how different compounds isolated from Xenia spp. and closely related corals could be explained from 7 using CYP and SHD chemistry. The oxidative cleavage of the cyclobutene ring in 7 has been proposed in the literature28. The end points of these putative biochemical pathways all represent isolated molecules from different Xenia spp8. and could all result from CYP and SDH chemistry following the outlined scheme. Acylations and methylations are also observed in some compounds but could be the result of non-clustered specific or promiscuous enzymatic activity. CYP = cytochrome P450 monooxygenase, IDS = isoprenyl diphosphate synthase, SDH = short-chain dehydrogenase, TC = terpene cyclase, MT = methyl transferase, AT = acyl transferase.
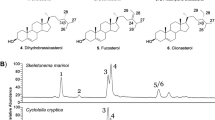
Extended Data Fig. 6 Analysis of a dichloromethane extract of Capnella imbricata by GC-MS.
Signals for capnellene (11), capnellene-8β,10α-diol (12) and putative terpenoid compounds are labeled.
Supplementary information
Supplementary Information
Supplementary Figs. 1–21, Notes 1 and 2, Tables 1–11 and references 1–51.
Rights and permissions
About this article
Cite this article
Burkhardt, I., de Rond, T., Chen, P.YT. et al. Ancient plant-like terpene biosynthesis in corals. Nat Chem Biol 18, 664–669 (2022). https://doi.org/10.1038/s41589-022-01026-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01026-2
This article is cited by
-
AlphaFold predictions are valuable hypotheses and accelerate but do not replace experimental structure determination
Nature Methods (2024)
-
Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase
Nature Communications (2023)
-
Coral–algal endosymbiosis characterized using RNAi and single-cell RNA-seq
Nature Microbiology (2023)
-
Design of a redox-proficient Escherichia coli for screening terpenoids and modifying cytochrome P450s
Nature Catalysis (2023)
-
Old path, new frontier
Nature Chemical Biology (2022)