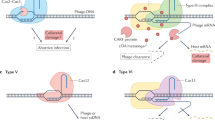
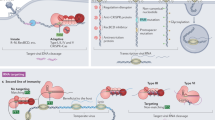
Abstract
CRISPR–Cas systems provide resistance against foreign mobile genetic elements and have a wide range of genome editing and biotechnological applications. In this Review, we examine recent advances in understanding the molecular structures and mechanisms of enzymes comprising bacterial RNA-guided CRISPR–Cas immune systems and deployed for wide-ranging genome editing applications. We explore the adaptive and interference aspects of CRISPR–Cas function as well as open questions about the molecular mechanisms responsible for genome targeting. These structural insights reflect close evolutionary links between CRISPR–Cas systems and mobile genetic elements, including the origins and evolution of CRISPR–Cas systems from DNA transposons, retrotransposons and toxin–antitoxin modules. We discuss how the evolution and structural diversity of CRISPR–Cas systems explain their functional complexity and utility as genome editing tools.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Knott, G. J. & Doudna, J. A. CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018).
Koonin, E. V. & Makarova, K. S. Origins and evolution of CRISPR-Cas systems. Phil. Trans. R. Soc. B 374, 20180087 (2019).
Hille, F. et al. The biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259 (2018).
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397 (2015).
Grissa, I., Vergnaud, G. & Pourcel, C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8, 172 (2007).
McGinn, J. & Marraffini, L. A. Molecular mechanisms of CRISPR–Cas spacer acquisition. Nat. Rev. Microbiol. 17, 7–12 (2019).
Gleditzsch, D. et al. PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 16, 504–517 (2019).
Behler, J. & Hess, W. R. Approaches to study CRISPR RNA biogenesis and the key players involved. Methods 172, 12–26 (2020).
Marino, N. D., Pinilla-Redondo, R., Csörgő, B. & Bondy-Denomy, J. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods 17, 471–479 (2020).
Makarova, K. S. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2019).
Jia, N. & Patel, D. J. Structure-based functional mechanisms and biotechnology applications of anti-CRISPR proteins. Nat. Rev. Mol. Cell Biol. 22, 563–579 (2021).
Liu, T. Y. & Doudna, J. A. Chemistry of class 1 CRISPR-Cas effectors: binding, editing, and regulation. J. Biol. Chem. 295, 14473–14487 (2020).
Molina, R., Sofos, N. & Montoya, G. Structural basis of CRISPR-Cas type III prokaryotic defence systems. Curr. Opin. Struct. Biol. 65, 119–129 (2020).
Taylor, H. N. et al. Positioning diverse type IV structures and functions within class 1 CRISPR-Cas systems. Front. Microbiol. 12, 674522 (2021).
O’Connell, M. R. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J. Mol. Biol. 431, 66–87 (2019).
Krupovic, M., Makarova, K. S., Forterre, P., Prangishvili, D. & Koonin, E. V. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 12, 36 (2014).
Hickman, A. B., Kailasan, S., Genzor, P., Haase, A. D. & Dyda, F. Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas. eLife 9, e50004 (2020). This work describes the structure of the casposase and its site specificity, providing insight into the evolutionary origins of the Cas1 protein.
Wang, J. et al. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163, 840–853 (2015).
Nuñez, J. K., Harrington, L. B., Kranzusch, P. J., Engelman, A. N. & Doudna, J. A. Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature 527, 535–538 (2015).
Xiao, Y., Ng, S., Nam, K. H. & Ke, A. How type II CRISPR–Cas establish immunity through Cas1–Cas2-mediated spacer integration. Nature 550, 137–141 (2017).
Wright, A. V. et al. Structures of the CRISPR genome integration complex. Science 357, 1113–1118 (2017).
Hickman, A. B. & Dyda, F. DNA transposition at work. Chem. Rev. 116, 12758–12784 (2016).
Béguin, P., Charpin, N., Koonin, E. V., Forterre, P. & Krupovic, M. Casposon integration shows strong target site preference and recapitulates protospacer integration by CRISPR-Cas systems. Nucleic Acids Res. 44, 10367–10376 (2016).
Yosef, I., Goren, M. G. & Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576 (2012).
Datsenko, K. A. et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 3, 945 (2012).
Arslan, Z., Hermanns, V., Wurm, R., Wagner, R. & Pul, Ü. Detection and characterization of spacer integration intermediates in type IE CRISPR–Cas system. Nucleic Acids Res. 42, 7884–7893 (2014).
Nuñez, J. K., Lee, A. S. Y., Engelman, A. & Doudna, J. A. Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature 519, 193–198 (2015).
Rollie, C., Schneider, S., Brinkmann, A. S., Bolt, E. L. & White, M. F. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. eLife 4, e08716 (2015).
Wright, A. V. & Doudna, J. A. Protecting genome integrity during CRISPR immune adaptation. Nat. Struct. Mol. Biol. 23, 876–883 (2016).
Nuñez, J. K. et al. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 21, 528–534 (2014).
Sasnauskas, G. & Siksnys, V. CRISPR adaptation from a structural perspective. Curr. Opin. Struct. Biol. 65, 17–25 (2020).
Wilkinson, M. et al. Structure of the DNA-bound spacer Capture complex of a type II CRISPR-Cas system. Mol. Cell 75, 90–101.e5 (2019).
Hu, C. et al. Mechanism for Cas4-assisted directional spacer acquisition in CRISPR–Cas. Nature 598, 515–520 (2021). This work provides the mechanism and structural basis for Cas4-assisted PAM processing and describes a model in which PAM sequestration and delayed processing influences the orientation of spacer integration.
Burstein, D. et al. New CRISPR–Cas systems from uncultivated microbes. Nature 542, 237–241 (2016).
Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J. 1, 325–336 (2018).
Wright, A. V. et al. A functional mini-integrase in a two-protein type V-C CRISPR system. Mol. Cell 73, 727–737 (2019). This work describes a tetrameric CRISPR integrase that may represent the ancestral CRISPR integrase.
Santiago-Frangos, A., Buyukyoruk, M., Wiegand, T., Krishna, P. & Wiedenheft, B. Distribution and phasing of sequence motifs that facilitate CRISPR adaptation. Curr. Biol. 31, 3515–3524 (2021).
Koonin, E. V. & Makarova, K. S. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 10, 679–686 (2013).
Bertelsen, M. B. et al. Structural basis for toxin inhibition in the VapXD toxin-antitoxin system. Structure 29, 139–150 (2021).
Kwon, A.-R. et al. Structural and biochemical characterization of HP0315 from Helicobacter pylori as a VapD protein with an endoribonuclease activity. Nucleic Acids Res. 40, 4216–4228 (2012).
Ka, D., Kim, D., Baek, G. & Bae, E. Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions. Biochem. Biophys. Res. Commun. 451, 152–157 (2014).
Silas, S. et al. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science 351, aad4234 (2016).
Koonin, E. V. & Makarova, K. S. Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. Genome Biol. Evol. 9, 2812–2825 (2017).
Silas, S. et al. On the origin of reverse transcriptase-using CRISPR-Cas systems and their hyperdiverse, enigmatic spacer repertoires. mBio 8, e00897-17 (2017).
Mohr, G. et al. A reverse transcriptase-Cas1 fusion protein contains a Cas6 domain required for both CRISPR RNA biogenesis and RNA spacer acquisition. Mol. Cell 72, 700–714 (2018).
Wang, J. Y. et al. Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex. Nat. Commun. 12, 2571 (2021). This work describes the structure of a Cas6–RT–Cas1–Cas2 complex, highlighting interactions between the three domains and the potential functional implications for CRISPR adaptation.
Stamos, J. L., Lentzsch, A. M. & Lambowitz, A. M. Structure of a thermostable group II intron reverse transcriptase with template-primer and its functional and evolutionary implications. Mol. Cell 68, 926–939 (2017).
Nussenzweig, P. M. & Marraffini, L. A. Molecular mechanisms of CRISPR-Cas immunity in bacteria. Annu. Rev. Genet. 54, 93–120 (2020).
Levy, A. et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510 (2015).
Wigley, D. B. Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 11, 9–13 (2013).
Kim, S. et al. Selective loading and processing of prespacers for precise CRISPR adaptation. Nature 579, 141–145 (2020). This work describes how the kinetic coordination of prespacer processing and PAM trimming affects the orientation of spacer integration and presents a model for prespacer selection and processing.
Ramachandran, A., Summerville, L., Learn, B. A., DeBell, L. & Bailey, S. Processing and integration of functionally oriented prespacers in the Escherichia coli CRISPR system depends on bacterial host exonucleases. J. Biol. Chem. 295, 3403–3414 (2020).
Kieper, S. N. et al. Cas4 facilitates PAM-compatible spacer selection during CRISPR adaptation. Cell Rep. 22, 3377–3384 (2018).
Lee, H., Dhingra, Y. & Sashital, D. G. The Cas4-Cas1-Cas2 complex mediates precise prespacer processing during CRISPR adaptation. eLife 8, e44248 (2019).
Musharova, O. et al. Prespacers formed during primed adaptation associate with the Cas1–Cas2 adaptation complex and the Cas3 interference nuclease–helicase. Proc. Natl Acad. Sci. USA 118, e2021291118 (2021).
Wu, C. et al. Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis. Nucleic Acids Res. 49, 2973–2984 (2021).
Drabavicius, G. et al. DnaQ exonuclease-like domain of Cas2 promotes spacer integration in a type I-E CRISPR-Cas system. EMBO Rep. 19, e45543 (2018).
Lee, H., Zhou, Y., Taylor, D. W. & Sashital, D. G. Cas4-dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol. Cell 70, 48–59.e5 (2018).
Heler, R. et al. Cas9 specifies functional viral targets during CRISPR–Cas adaptation. Nature 519, 199–202 (2015).
Heler, R. et al. Mutations in Cas9 enhance the rate of acquisition of viral spacer sequences during the CRISPR-Cas immune response. Mol. Cell 65, 168–175 (2017).
Jakhanwal, S. et al. A CRISPR-Cas9–integrase complex generates precise DNA fragments for genome integration. Nucleic Acids Res. 49, 3546–3556 (2021).
Swarts, D. C., Mosterd, C., van Passel, M. W. J. & Brouns, S. J. J. CRISPR interference directs strand specific spacer acquisition. PLoS ONE 7, e35888 (2012).
Dillard, K. E. et al. Assembly and translocation of a CRISPR-Cas primed acquisition complex. Cell 175, 934–946.e15 (2018).
Xue, C., Whitis, N. R. & Sashital, D. G. Conformational control of Cascade interference and priming activities in CRISPR immunity. Mol. Cell 64, 826–834 (2016).
Nicholson, T. J. et al. Bioinformatic evidence of widespread priming in type I and II CRISPR-Cas systems. RNA Biol. 16, 566–576 (2019).
Nussenzweig, P. M., McGinn, J. & Marraffini, L. A. Cas9 cleavage of viral genomes primes the acquisition of new immunological memories. Cell Host Microbe 26, 515–526.e6 (2019).
Shipman, S. L., Nivala, J., Macklis, J. D. & Church, G. M. Molecular recordings by directed CRISPR spacer acquisition. Science 353, aaf1175 (2016).
Sheth, R. U., Yim, S. S., Wu, F. L. & Wang, H. H. Multiplex recording of cellular events over time on CRISPR biological tape. Science 358, 1457–1461 (2017).
Schmidt, F., Cherepkova, M. Y. & Platt, R. J. Transcriptional recording by CRISPR spacer acquisition from RNA. Nature 562, 380–385 (2018).
Munck, C., Sheth, R. U., Freedberg, D. E. & Wang, H. H. Recording mobile DNA in the gut microbiota using an Escherichia coli CRISPR-Cas spacer acquisition platform. Nat. Commun. 11, 95 (2020).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Gasiunas, G. & Barrangou, R. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA 109, E2579–E2586 (2012).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Hwang, W. Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013).
Cho, S. W., Kim, S., Kim, J. M. & Kim, J.-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232 (2013).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Zetsche, B. et al. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34 (2017).
Liu, J.-J. et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 566, 218–223 (2019).
Strecker, J. et al. Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 10, 212 (2019).
Pausch, P. et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369, 333–337 (2020).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Swarts, D. C. & Jinek, M. Cas9 versus Cas12a/Cpf1: structure-function comparisons and implications for genome editing. Wiley Interdiscip. Rev. RNA 9, e1481 (2018).
Stella, S., Alcón, P. & Montoya, G. Class 2 CRISPR–Cas RNA-guided endonucleases: Swiss Army knives of genome editing. Nat. Struct. Mol. Biol. 24, 882–892 (2017).
Altae-Tran, H. et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65 (2021).
Jinek, M. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997 (2014).
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014). This study reveals for the first time how Cas9 recognizes DNA.
Jiang, F., Zhou, K., Ma, L., Gressel, S. & Doudna, J. A. A Cas9–guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481 (2015).
Anders, C., Niewoehner, O., Duerst, A. & Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014).
Jiang, F. et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 (2016).
Sternberg, S. H., LaFrance, B., Kaplan, M. & Doudna, J. A. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–113 (2015).
Zhang, Y. et al. Catalytic-state structure and engineering of Streptococcus thermophilus Cas9. Nat. Catal. 3, 813–823 (2020).
Sun, W. et al. Structures of Neisseria meningitidis Cas9 complexes in catalytically poised and anti-CRISPR-inhibited states. Mol. Cell 76, 938–952.e5 (2019).
Pacesa, M. & Jinek, M. Mechanism of R-loop formation and conformational activation of Cas9. Preprint at bioRxiv https://doi.org/10.1101/2021.09.16.460614 (2021).
Bravo, J. P. K. et al. Structural basis for mismatch surveillance by CRISPR-Cas9. Nature 603, 343–347 (2022). This study demonstrates how excessive target mismatches inhibit DNA cutting by Cas9 and reveals a most comprehensive structure of Cas9 bound to the DNA cleavage product.
Zhu, X. et al. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat. Struct. Mol. Biol. 26, 679–685 (2019).
Chen, J. S. et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Palermo, G. et al. Protospacer adjacent motif-induced allostery activates CRISPR-Cas9. J. Am. Chem. Soc. 139, 16028–16031 (2017).
Palermo, G. et al. Key role of the REC lobe during CRISPR-Cas9 activation by ‘sensing’, ‘regulating’, and ‘locking’ the catalytic HNH domain. Q. Rev. Biophys. 51, e91 (2018).
Nierzwicki, L. et al. Enhanced specificity mutations perturb allosteric signaling in CRISPR-Cas9. eLife 10, e73601 (2021).
Zuo, Z. et al. Structural and functional insights into the bona fide catalytic state of Streptococcus pyogenes Cas9 HNH nuclease domain. eLife 8, e46500 (2019).
Belato, H. B. et al. Structural and dynamic insights into the HNH nuclease of divergent Cas9 species. J. Struct. Biol. 214, 107814 (2021).
Globyte, V., Lee, S. H., Bae, T., Kim, J. & Joo, C. CRISPR /Cas9 searches for a protospacer adjacent motif by lateral diffusion. EMBO J. 38, e99466 (2019).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014).
Cofsky, J. C., Soczek, K. M., Knott, G. J., Nogales, E. & Doudna, J. A. CRISPR-Cas9 bends and twists DNA to read its sequence. Nat. Struct. Mol. Biol. 29, 395–402 (2022). This article for the first time reveals structural insights into how Cas9 opens dsDNA to interrogate target sequences.
Ivanov, I. E. et al. Cas9 interrogates DNA in discrete steps modulated by mismatches and supercoiling. Proc. Natl Acad. Sci. USA 117, 5853–5860 (2020).
Liu, M.-S. et al. Engineered CRISPR/Cas9 enzymes improve discrimination by slowing DNA cleavage to allow release of off-target DNA. Nat. Commun. 11, 3576 (2020).
Kleinstiver, B. P. et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Yamano, T. et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962 (2016). This study reveals for the first time how Cas12a recognizes DNA.
Dong, D. et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522–526 (2016).
Pausch, P. et al. DNA interference states of the hypercompact CRISPR-CasΦ effector. Nat. Struct. Mol. Biol. 28, 652–661 (2021). This article describes how a minimal Cas12 enzyme binds dsDNA and catalyses DNA cleavage.
Carabias, A. et al. Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3. Nat. Commun. 12, 4476 (2021).
Yang, H., Gao, P., Rajashankar, K. R. & Patel, D. J. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell 167, 1814–1828 (2016).
Tsuchida, C. A. et al. Chimeric CRISPR-CasX enzymes and guide RNAs for improved genome editing activity. Mol. Cell 82, 1199–1209.e6 (2022).
Harrington, L. B. et al. A scoutRNA is required for some type V CRISPR-Cas systems. Mol. Cell 79, 416–424.e5 (2020).
Huang, C. J., Adler, B. A. & Doudna, J. A. A naturally DNase-free CRISPR-Cas12c enzyme silences gene expression. Preprint at bioRxiv https://doi.org/10.1101/2021.12.06.471469 (2021).
Kurihara, N. et al. Structure of the type V-C CRISPR-Cas effector enzyme. Mol. Cell 82, 1–13 (2022).
Harrington, L. B. et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842 (2018).
Takeda, S. N. et al. Structure of the miniature type V-F CRISPR-Cas effector enzyme. Mol. Cell 81, 558–570 (2021).
Xiao, R., Li, Z., Wang, S., Han, R. & Chang, L. Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR–Cas12f nuclease. Nucleic Acids Res. 49, 4120–4128 (2021).
Li, Z., Zhang, H., Xiao, R., Han, R. & Chang, L. Cryo-EM structure of the RNA-guided ribonuclease Cas12g. Nat. Chem. Biol. 17, 387–393 (2021).
Swarts, D. C., van der Oost, J. & Jinek, M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell 66, 221–233 (2017).
Gao, P., Yang, H., Rajashankar, K. R., Huang, Z. & Patel, D. J. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 26, 901–913 (2016).
Stella, S., Alcón, P. & Montoya, G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature 546, 559–563 (2017).
Cofsky, J. C. et al. CRISPR-Cas12a exploits R-loop asymmetry to form double-strand breaks. eLife 9, e55143 (2020).
Stella, S. et al. Conformational activation promotes CRISPR-Cas12a catalysis and resetting of the endonuclease activity. Cell 175, 1856–1871 (2018). This study provides extensive structural and mechanistic insights into the conformational activation of Cas12a.
Huang, X. et al. Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2. Nat. Commun. 11, 5241 (2020).
Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
Swarts, D. C. & Jinek, M. Mechanistic insights into the cis- and trans-acting DNase activities of Cas12a. Mol. Cell 73, 589–600 (2019).
Jiang, W. et al. CRISPR-Cas12a nucleases bind flexible DNA duplexes without RNA/DNA complementarity. ACS Omega 4, 17140–17147 (2019).
Paul, B., Chaubet, L., Verver, D. E. & Montoya, G. Mechanics of CRISPR-Cas12a and engineered variants on λ-DNA. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab1272 (2021).
Losito, M., Smith, Q. M., Newton, M. D., Cuomo, M. E. & Rueda, D. S. Cas12a target search and cleavage on force-stretched DNA. Phys. Chem. Chem. Phys. 23, 26640–26644 (2021).
Kapitonov, V. V., Makarova, K. S. & Koonin, E. V. ISC, a novel group of bacterial and archaeal DNA transposons that encode Cas9 homologs. J. Bacteriol. 198, 797–807 (2015).
Shmakov, S. et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 15, 169–182 (2017).
Karvelis, T. et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696 (2021).
Weinberg, Z., Perreault, J., Meyer, M. M. & Breaker, R. R. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462, 656–659 (2009).
Stoddard, B. L. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob. DNA 5, 7 (2014).
Stoddard, B. L. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure 19, 7–15 (2011).
Klompe, S. E., Vo, P. L. H., Halpin-Healy, T. S. & Sternberg, S. H. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019).
Strecker, J. et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019).
Faure, G. et al. CRISPR–Cas in mobile genetic elements: counter-defence and beyond. Nat. Rev. Microbiol. 17, 513–525 (2019).
Peters, J. E., Makarova, K. S., Shmakov, S. & Koonin, E. V. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc. Natl Acad. Sci. USA 114, E7358–E7366 (2017).
Rybarski, J. R., Hu, K., Hill, A. M., Wilke, C. O. & Finkelstein, I. J. Metagenomic discovery of CRISPR-associated transposons. Proc. Natl Acad. Sci. USA 118, e2112279118 (2021).
Halpin-Healy, T. S., Klompe, S. E., Sternberg, S. H. & Fernández, I. S. Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system. Nature 577, 271–274 (2020). This work provides mechanistic insights into subtype I-F3 CRISPR transposases by describing the structures of a TniQ–Cascade complex and reveals interactions between TniQ and Cas6 and Cas7.1 within the Cascade complex.
Li, Z., Zhang, H., Xiao, R. & Chang, L. Cryo-EM structure of a type I-F CRISPR RNA guided surveillance complex bound to transposition protein TniQ. Cell Res. 30, 179–181 (2020).
Jia, N., Xie, W., de la Cruz, M. J., Eng, E. T. & Patel, D. J. Structure-function insights into the initial step of DNA integration by a CRISPR-Cas-Transposon complex. Cell Res. 30, 182–184 (2020).
Wang, B., Xu, W. & Yang, H. Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex. Cell Res. 30, 185–187 (2020).
Park, J.-U. et al. Structural basis for target site selection in RNA-guided DNA transposition systems. Science 373, 768–774 (2021). This work provides mechanistic insights into subtype V-K CRISPR transposases by describing a transposition regulator, TnsC, from a subtype V-K CAST system and its interaction with TniQ and proposing a model for subtype V-K CAST transposition.
Querques, I., Schmitz, M., Oberli, S., Chanez, C. & Jinek, M. Target site selection and remodelling by type V CRISPR-transposon systems. Nature 599, 497–502 (2021). This work provides mechanistic insights into subtype V-K CRISPR transposases by describing target recognition by Cas12k and the role of the transposition regulator TnsC and proposing an alternative model for subtype V-K CAST transposition.
Xiao, R. et al. Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition. Mol. Cell 81, 4457–4466.e5 (2021).
Chowdhury, S. et al. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell 169, 47–57.e11 (2017).
Guo, T. W. et al. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell 171, 414–426.e12 (2017).
Pausch, P. et al. Structural variation of type I-F CRISPR RNA guided DNA surveillance. Mol. Cell 67, 622–632.e4 (2017).
Rollins, M. F. et al. Structure reveals a mechanism of CRISPR-RNA-guided nuclease recruitment and anti-CRISPR viral mimicry. Mol. Cell 74, 132–142.e5 (2019).
Hayes, R. P. et al. Structural basis for promiscuous PAM recognition in type I–E Cascade from E. coli. Nature 530, 499–503 (2016).
Greene, E. C. & Mizuuchi, K. Dynamics of a protein polymer: the assembly and disassembly pathways of the MuB transposition target complex. EMBO J. 21, 1477–1486 (2002).
Vo, P. L. H. et al. CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol. 39, 480–489 (2021).
Rubin, B. E. et al. Species- and site-specific genome editing in complex bacterial communities. Nat. Microbiol. 7, 34–47 (2021).
Stellwagen, A. E. & Craig, N. L. Avoiding self: two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 16, 6823–6834 (1997).
Saito, M. et al. Dual modes of CRISPR-associated transposon homing. Cell 184, 2441–2453 (2021).
Petassi, M. T., Hsieh, S.-C. & Peters, J. E. Guide RNA categorization enables target site choice in Tn7-CRISPR-Cas transposons. Cell 183, 1757–1771.e18 (2020).
Waddell, C. S. & Craig, N. L. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 2, 137–149 (1988).
Klompe, S. E. et al. Evolutionary and mechanistic diversity of type I-F CRISPR-associated transposons. Mol. Cell 82, 616–628 (2022).
Liu, G., Lin, Q., Jin, S. & Gao, C. The CRISPR-Cas toolbox and gene editing technologies. Mol. Cell 82, 333–347 (2022).
Nambiar, T. S., Baudrier, L., Billon, P. & Ciccia, A. CRISPR-based genome editing through the lens of DNA repair. Mol. Cell 82, 348–388 (2022).
Lapinaite, A. et al. DNA capture by a CRISPR-Cas9 guided adenine base editor. Science 369, 566–571 (2022).
Hirano, S., Nishimasu, H., Ishitani, R. & Nureki, O. Structural basis for the altered PAM specificities of engineered CRISPR-Cas9. Mol. Cell 61, 886–894 (2016).
Chen, W. et al. Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease. PLoS Biol. 17, e3000496 (2019).
Anders, C., Bargsten, K. & Jinek, M. Structural plasticity of PAM recognition by engineered variants of the RNA-guided endonuclease Cas9. Mol. Cell 61, 895–902 (2016).
Guo, M. et al. Structural insights into a high fidelity variant of SpCas9. Cell Res. 29, 183–192 (2019).
Nishimasu, H. et al. Structural basis for the altered PAM recognition by engineered CRISPR-Cpf1. Mol. Cell 67, 139–147.e2 (2017).
Shams, A. et al. Comprehensive deletion landscape of CRISPR-Cas9 identifies minimal RNA-guided DNA-binding modules. Nat. Commun. 12, 5664 (2021).
Donohoue, P. D. et al. Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells. Mol. Cell 81, 3637–3649.e5 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Townshend, R. J. L. et al. Geometric deep learning of RNA structure. Science 373, 1047–1051 (2021).
Wei, J., Chen, S., Zong, L., Gao, X. & Li, Y. Protein-RNA interaction prediction with deep learning: structure matters. Preprint at arXiv https://arxiv.org/abs/2107.12243 (2021).
Nierzwicki, Ł. & Palermo, G. Molecular dynamics to predict cryo-EM: capturing transitions and short-lived conformational states of biomolecules. Front. Mol. Biosci. 8, 641208 (2021).
Wang, J. et al. Gaussian accelerated molecular dynamics: principles and applications. Wiley Interdiscip. Rev. Comput. Mol. Sci. 11, e1521 (2021).
Xiao, Y., Luo, M., Dolan, A. E., Liao, M. & Ke, A. Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science 361, eaat0839 (2018).
Acknowledgements
The authors thank J. C. Cofsky, K. M. Soczek and G. J. Knott for sharing the coordinates of Cas9 bound to linear and bent double-stranded DNA before publication. J.Y.W. is supported by the US National Science Foundation Graduate Fellowship and was previously supported by the Berkeley Graduate Fellowship. P.P. receives funding from the European Regional Development Fund under grant agreement number 01.2.2-CPVA-V-716-01-0001 with the Central Project Management Agency (CPVA), Lithuania, and from the Research Council of Lithuania (LMTLT) under grant agreement number S-MIP-22-10. This material is based upon work supported by the US National Science Foundation under award number 1817593 and by the Somatic Cell Genome Editing Program of the Common Fund of the US National Institutes of Health under award number U01AI142817-02. J.A.D. is a Howard Hughes Medical Institute investigator. The authors thank G. J. Knott and A. Lapinaite for helpful discussions.
Author information
Authors and Affiliations
Contributions
All authors contributed to writing, reviewing and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.A.D. is a cofounder of Caribou Biosciences, Editas Medicine, Scribe Therapeutics, Intellia Therapeutics and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Vertex, Caribou Biosciences, Intellia Therapeutics, Scribe Therapeutics, Mammoth Biosciences, Synthego, Algen Biotechnologies, Felix Biosciences, The Column Group and Inari. J.A.D. is the Chief Science Advisor of Sixth Street, is on the Board of Directors at Altos, Johnson & Johnson and Tempus, and has research projects sponsored by Biogen, Pfizer, AppleTree Partners and Roche. The Regents of the University of California have patents issued and pending for CRISPR technologies on which P.P. and J.A.D. are named as inventors. J.Y.W. declares no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Hong Li, Guillermo Montoya and Dinshaw Patel for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, J.Y., Pausch, P. & Doudna, J.A. Structural biology of CRISPR–Cas immunity and genome editing enzymes. Nat Rev Microbiol 20, 641–656 (2022). https://doi.org/10.1038/s41579-022-00739-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-022-00739-4
This article is cited by
-
An alpha-helical lid guides the target DNA toward catalysis in CRISPR-Cas12a
Nature Communications (2024)
-
Good reasons for structural biology
Nature Structural & Molecular Biology (2024)
-
CRISPR technologies for genome, epigenome and transcriptome editing
Nature Reviews Molecular Cell Biology (2024)
-
Structural basis of Gabija anti-phage defence and viral immune evasion
Nature (2024)
-
A birds-eye-view on CRISPR-Cas system in agriculture
The Nucleus (2024)