Abstract
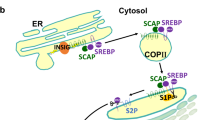
Tumorigenesis is associated with elevated glucose and glutamine consumption, but how cancer cells can sense their levels to activate lipid synthesis is unknown. Here, we reveal that ammonia, released from glutamine, promotes lipogenesis via activation of sterol regulatory element-binding proteins (SREBPs), endoplasmic reticulum-bound transcription factors that play a central role in lipid metabolism. Ammonia activates the dissociation of glucose-regulated, N-glycosylated SREBP-cleavage-activating protein (SCAP) from insulin-inducible gene protein (Insig), an endoplasmic reticulum-retention protein, leading to SREBP translocation and lipogenic gene expression. Notably, 25-hydroxycholesterol blocks ammonia to access its binding site on SCAP. Mutating aspartate D428 to alanine prevents ammonia binding to SCAP, abolishes SREBP-1 activation and suppresses tumour growth. Our study characterizes the unknown role, opposite to sterols, of ammonia as a key activator that stimulates SCAP–Insig dissociation and SREBP-1 activation to promote tumour growth and demonstrates that SCAP is a critical sensor of glutamine, glucose and sterol levels to precisely control lipid synthesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data that support the findings of this study are available within the paper and its supplementary information files. RNA-seq data for Figs. 1a and 2f and Extended Data Fig. 1a are provided in Supplementary Data and the raw data are deposited in the Gene Expression Omnibus (accession no. GSE199089). Source data are provided with this paper.
Code availability
No custom codes were used during this study.
References
Guo, D., Bell, E. H., Mischel, P. & Chakravarti, A. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr. Pharm. Des. 20, 2619–2626 (2014).
Cheng, C., Geng, F., Cheng, X. & Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 38, 27 (2018).
Cheng, X., Li, J. & Guo, D. SCAP/SREBPs are central players in lipid metabolism and novel metabolic targets in cancer therapy. Curr. Top. Med. Chem. 18, 484–493 (2018).
Nohturfft, A. & Zhang, S. C. Coordination of lipid metabolism in membrane biogenesis. Annu. Rev. Cell Dev. Biol. 25, 539–566 (2009).
Goldstein, J. L., DeBose-Boyd, R. A. & Brown, M. S. Protein sensors for membrane sterols. Cell 124, 35–46 (2006).
Goldstein, J. L. & Brown, M. S. A century of cholesterol and coronaries: from plaques to genes to statins. Cell 161, 161–172 (2015).
Jeon, T. I. & Osborne, T. F. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23, 65–72 (2012).
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002).
Brown, M. S., Radhakrishnan, A. & Goldstein, J. L. Retrospective on cholesterol homeostasis: the central role of scap. Annu. Rev. Biochem. 87, 783–807 (2018).
Bennett, M. K., Lopez, J. M., Sanchez, H. B. & Osborne, T. F. Sterol regulation of fatty acid synthase promoter. Coordinate feedback regulation of two major lipid pathways. J. Biol. Chem. 270, 25578–25583 (1995).
Xu, D. et al. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 580, 530–535 (2020).
Guo, D. et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci. Signal 2, ra82 (2009).
Guo, D. et al. An LXR agonist promotes GBM cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov. 1, 442–456 (2011).
Li, C. et al. SREBP-1 has a prognostic role and contributes to invasion and metastasis in human hepatocellular carcinoma. Int. J. Mol. Sci. 15, 7124–7138 (2014).
Zhu, Z. et al. p54(nrb)/NONO regulates lipid metabolism and breast cancer growth through SREBP-1A. Oncogene 35, 1399–1410 (2016).
Sun, Y. et al. Role of transgelin-2 in diabetes-associated pancreatic ductal adenocarcinoma. Oncotarget 8, 49592–49604 (2017).
Gao, Y. et al. SREBP1 promotes the invasion of colorectal cancer accompanied upregulation of MMP7 expression and NF-κB pathway activation. BMC Cancer 19, 685 (2019).
Sun, L. P., Seemann, J., Goldstein, J. L. & Brown, M. S. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in SCAP inaccessible to COPII proteins. Proc. Natl. Acad. Sci. USA 104, 6519–6526 (2007).
Espenshade, P. J., Cheng, D., Goldstein, J. L. & Brown, M. S. Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274, 22795–22804 (1999).
Cheng, D. et al. Secreted site-1 protease cleaves peptides corresponding to luminal loop of sterol regulatory element-binding proteins. J. Biol. Chem. 274, 22805–22812 (1999).
Rawson, R. B. et al. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1, 47–57 (1997).
Duncan, E. A., Dave, U. P., Sakai, J., Goldstein, J. L. & Brown, M. S. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J. Biol. Chem. 273, 17801–17809 (1998).
Hua, X., Sakai, J., Brown, M. S. & Goldstein, J. L. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J. Biol. Chem. 271, 10379–10384 (1996).
Yang, T. et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110, 489–500 (2002).
Yabe, D., Brown, M. S. & Goldstein, J. L. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. USA 99, 12753–12758 (2002).
Espenshade, P. J., Li, W. P. & Yabe, D. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. USA 99, 11694–11699 (2002).
Adams, C. M., Goldstein, J. L. & Brown, M. S. Cholesterol-induced conformational change in SCAP enhanced by Insig proteins and mimicked by cationic amphiphiles. Proc. Natl. Acad. Sci. USA 100, 10647–10652 (2003).
Cheng, C. et al. Glucose-mediated N-glycosylation of SCAP is essential for SREBP-1 activation and tumor growth. Cancer Cell 28, 569–581 (2015).
Guo, D. SCAP links glucose to lipid metabolism in cancer cells. Mol. Cell. Oncol. https://doi.org/10.1080/23723556.2015.1132120 (2016).
Shao, W. & Espenshade, P. J. Sugar makes fat by talking to SCAP. Cancer Cell 28, 548–549 (2015).
Cheng, C. et al. Analysis of SCAP N-glycosylation and trafficking in human cells. J. Vis. Exp. https://doi.org/10.3791/54709 (2016).
Yang, L., Venneti, S. & Nagrath, D. Glutaminolysis: a hallmark of cancer metabolism. Annu. Rev. Biomed. Eng. 19, 163–194 (2017).
Pavlova, N. N. & Thompson, C. B. The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016).
Hensley, C. T., Wasti, A. T. & DeBerardinis, R. J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Invest. 123, 3678–3684 (2013).
Cheng, X., Geng, F. & Guo, D. DGAT1 protects tumor from lipotoxicity, emerging as a promising metabolic target for cancer therapy. Mol. Cell Oncol. 7, 1805257 (2020).
Wu, X. et al. Lipid droplets maintain energy homeostasis and glioblastoma growth via autophagic release of stored fatty acids. iScience 23, 101569 (2020).
Venneti, S. et al. Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci. Transl. Med. 7, 274ra217 (2015).
Reinfeld, B. I. et al. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature 593, 282–288 (2021).
Altman, B. J., Stine, Z. E. & Dang, C. V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 749 (2016).
Tanaka, K. et al. Compensatory glutamine metabolism promotes glioblastoma resistance to mTOR inhibitor treatment. J. Clin. Invest. 125, 1591–1602 (2015).
Grkovski, M. et al. Pharmacokinetic assessment of (18)F-(2S,4R)-4-fluoroglutamine in patients with cancer. J. Nucl. Med. 61, 357–366 (2020).
Dunphy, M. P. S. et al. In vivo PET assay of tumor glutamine flux and metabolism: in-human trial of (18)F-(2S,4R)-4-fluoroglutamine. Radiology 287, 667–675 (2018).
McBrayer, S. K. et al. Transaminase inhibition by 2-hydroxyglutarate impairs glutamate biosynthesis and redox homeostasis in glioma. Cell 175, 101–116 e125 (2018).
Curthoys, N. P. & Watford, m. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 15, 133–159 (1995).
Li, T. & Le, A. Glutamine metabolism in cancer. Adv. Exp. Med. Biol. 1063, 13–32 (2018).
Eng, C. H., Yu, K., Lucas, J., White, E. & Abraham, R. T. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci. Signal 3, ra31 (2010).
Liu, Y. et al. The role of ASCT2 in cancer: a review. Eur. J. Pharmacol. 837, 81–87 (2018).
Nohturfft, A., Yabe, D., Goldstein, J. L., Brown, M. S. & Espenshade, P. J. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 102, 315–323 (2000).
Ghanakota, P. & Carlson, H. A. Driving structure-based drug discovery through cosolvent molecular dynamics. J. Med. Chem. 59, 10383–10399 (2016).
Yan, R. et al. A structure of human SCAP bound to Insig-2 suggests how their interaction is regulated by sterols. Science https://doi.org/10.1126/science.abb2224 (2021).
Kober, D. L. et al. SCAP structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing. Cell https://doi.org/10.1016/j.cell.2021.05.019 (2021).
Brown, M. S. & Goldstein, J. L. Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J. Lipid Res. 50, S15–S27 (2009).
Geng, F. et al. Inhibition of SOAT1 suppresses glioblastoma growth via blocking SREBP-1-mediated lipogenesis. Clin. Cancer Res. 22, 5337–5348 (2016).
Cheng, X. et al. Targeting DGAT1 ameliorates glioblastoma by increasing fat catabolism and oxidative stress. Cell Metab. 32, 229–242 (2020).
Ru, P. et al. Feedback loop regulation of SCAP/SREBP-1 by miR-29 modulates EGFR signaling-driven glioblastoma growth. Cell Rep. 16, 1527–1535 (2016).
Walker, V. Ammonia metabolism and hyperammonemic disorders. Adv. Clin. Chem. 67, 73–150 (2014).
Braissant, O., McLin, V. A. & Cudalbu, C. Ammonia toxicity to the brain. J. Inherit. Metab. Dis. 36, 595–612 (2013).
Kim, J. et al. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature 546, 168–172 (2017).
Spinelli, J. B. et al. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science 358, 941–946 (2017).
Feramisco, J. D. et al. Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. Proc. Natl. Acad. Sci. USA 102, 3242–3247 (2005).
Webb, B. & Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinforma. 54, 5.6.1–5.6.37 (2016).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Wu, E. L. et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Zoete, V., Cuendet, M. A., Grosdidier, A. & Michielin, O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 32, 2359–2368 (2011).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013).
Vanommeslaeghe, K. & MacKerell, A. D. Jr. Automation of the CHARMM general force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012).
Vanommeslaeghe, K. et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–38 (1996).
Acknowledgements
This work was supported by the National Institute of Neurological Disorders and Stroke and the National Cancer Institute (USA) grants NS104332, NS112935 and R01CA240726 to D.G., CA227874 to D.G. and A.C. and an American Cancer Society Research Scholar Grant RSG-14-228-01–CSM to D.G. We also appreciate the support from the OSU Comprehensive Cancer Center–Pelotonia Idea grant and Urban & Shelly Meyer Fund for Cancer Research to D.G. The authors thank M. Torres for editorial assistance.
Author information
Authors and Affiliations
Contributions
D.G. conceived the ideas. C.C. and D.G. designed the experiments. C.C., F.G., Y. Zhong, H.W. and X.C. performed the experiments. Z.L. and X.-l.C. conducted computational simulations, Y. Zhao and X.M. performed RNA-seq data and pathway analyses. C.C., F.G., A.C. and D.G. analyzed the data. C.H. and W.D. provided glioma and lung cancer TMA. C.C., Z.L., X.-l.C. and D.G. wrote the manuscript and all authors reviewed and approved the manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Jiyeon Kim, Evripidis Gavathiotis and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary handling editor: Alfredo Giménez-Cassina
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Glutamine activates SREBP-1 to promote cell proliferation.
a, Heat map comparison of metabolic and overall pathways based on RNA-seq data from H1299 cells under glucose, glutamine or a combination of glucose and glutamine vs. both free conditions (12 h) using the bioinformatics Ingenuity Pathway Analysis (IPA). #NUM, no activity pattern available. b, c, Western blot analysis of cell lysates of cells stimulated with glutamine for 12 h (b) or with 4 mM glutamine at the indicated times (c) under serum-free conditions (glucose 5 mM). d, Lipids derived from 14C-labeled glucose (0.5 μCi, 2 h) in cells after culturing cells with/without glutamine (4 mM) for 12 h in serum-free medium containing 5 mM non-labeled glucose. The results are presented as mean ± SEM (n = 3). e, Proliferation of cancer cells cultured in medium supplemented with 1% dialyzed FBS with/without glutamine (4 mM) or glucose (5 mM) (mean ± SD, n = 3). f, g, Western blot analysis of cells after infection with shRNA-expressing lentivirus for 48 h and then placed in fresh medium (5 mM glucose) with/without glutamine (4 mM) for another 12 h (left panels). Cell proliferation was determined under 1% dialyzed FBS (right panels). The results are shown as mean ± SD (n = 3). h, Western blot analysis of cells after treatment with atorvastatin (5 μM) for 12 h in 5% lipoprotein-deficient serum (LPDS) containing 5 mM glucose with/without glutamine (4 mM). i, Western blot analysis of cells after stimulation with EGF (20 ng/ml) for 12 h in serum-free medium (5 mM glucose) with/without glutamine (4 mM). j, Western blot analysis of cells after incubation with/without aspartate (0.15 mM), asparagine (0.38 mM), leucine (0.38 mM), methionine (0.1 mM), threonine (0.17 mM) or glutamine (2 mM) for 12 h in HBSS buffer (containing 5.6 mM glucose) supplemented with essential amino acids. The dose selected for each amino acid is same as their concentration included in RPMI 1640 medium. Significance was determined by unpaired and two-tailed Student’s t-test (d) or two-way ANOVA with Dunnett’s (e) or Tukey’s (g) multiple comparisons adjustment.
Extended Data Fig. 2 Ammonia activates SREBPs and lipogenesis.
a, Western blot analysis of cells stimulated with glutamine, NH4Cl, NH3·H2O, NaCl, NaOH or NaNO3 (all 4 mM) under serum-free medium (5 mM glucose) for 12 hr. b, Western blot analysis of cells stimulated with NaCl (12 hr) in the absence of glutamine under serum-free culture conditions containing 5 mM glucose. c, Representative IF images of cells after stimulation with glutamate (4 mM), α-KG (4 mM), lactate (10 mM) or glutamine (4 mM) for 12 hr under serum-free culture conditions (5 mM glucose). Scale bars, 10 μm. The nuclear intensity of SREBP-1 (bottom panel) was quantified over 30 cells by ImageJ (mean ± SEM, n ≥ 30). d–f, Western blot analysis of H1299 cells stimulated with glutamine (4 mM), glutamate (Glu, 4 mM), α-KG (4 mM), octyl-α-KG (OA-KG) (2 mM) or NH4Cl (4 mM) for 12 hr under serum-free culture conditions (5 mM glucose) (d). The levels of glutamate (e) and α-KG (f) in the cells were measured using the appropriate assay kits. The results (e and f) are presented as mean ± SEM (n = 3). g, Western blot analysis of H1299 cells stimulated with glutamine (4 mM) or NH4Cl (4 mM) for 12 hr in the presence of glucose (5 mM) after ATG5 siRNA knockdown for 24 hr. h, Western blot analysis of cells stimulated with NH4Cl at the indicated doses for 12 hr under serum-free culture conditions (5 mM glucose). i–k, Western blot analysis of membranes (for GFP-SCAP, PDI and SREBP precursors) and nuclear extracts (for N-terminal SREBPs and Lamin A) from HEK293T cells transfected with GFP (2 μg), GFP-SCAP wild-type (NNN) (2 μg) or its mutant QQQ (5 μg), obtained by replacing all three N-glycosylation residues asparagine (N) to glutamine (Q), together with full length Flag-SREBP-1a (i), -1c (j), or HA-SREBP -2 (k) for 24 hr and then stimulated with glutamine or NH4Cl (all 4 mM) for another 12 hr under serum-free culture conditions (5 mM glucose). Significance was determined by unpaired and two-tailed Student t-test or one-way ANOVA with Dunnett’s multiple comparisons adjustment.
Extended Data Fig. 3 Suppressing ammonia release from glutamine inhibits SREBPs.
a, Relative metabolite levels in H1299 cells after treatment with GPNA (5 mM) or CB-839 (100 nM) for 12 hr under serum-free medium containing glutamine (4 mM) and glucose (5 mM) via using appropriate assay kits (mean ± SEM, n = 3). Cell culture conditions upon treatment are the same for the subsequent panels. b, Relative glutamine consumption of cells treating with GPNA (5 mM) or CB-839 (100 nM) for 12 hr (mean ± SEM, n = 3). c, d, Western blot analysis of cells treated with GPNA or CB-839 (48 hr). e, Western blot analysis of GBM30 cells treated with CB-839 (200 nM) for 12 hr with/without glutamine, glutamate or NH4Cl (all 4 mM). f, Ammonia measurement (left panel) in tumour tissues from H1299 cells (4 × 106) derived xenograft model treated with CB-839 (30 mg/kg/mouse, i.p., twice per day for 3 days) when tumour size reached 200 mm3 (mean ± SEM, n = 6). Middle panel shows representative IHC images. Scale bars, 50 μm. The expression levels were quantified by using ImageJ to analyze 4 images per tumour (3 tumours/group) (mean ± SEM, n ≥ 2441 cells) (right panel). g, Relative glutamine consumption (12 hr) of cells in culture condition as (a) after infection with shRNA-expressing lentiviruses (48 hr) (mean ± SEM, n = 3). h, Relative metabolite levels (12 hr) in cells in culture condition as (a) measured by the appropriate assay kit after infection with shRNA-expressing lentiviruses (48 hr) (mean ± SD, (n = 3). i, j, Western blot analysis of cells after infection with shRNA-expressing lentiviruses for 48 hr and then stimulated with 4 mM glutamate, α-KG or NH4Cl for 12 hr. k, l, Real-time qPCR (k) and Western blot (l) analysis of cells under serum-free medium containing 5 mM glucose and 4 mM glutamine for 12 hr after siRNA knockdown of glutamate dehydrogenase (GDH1/2), asparaginase (ASPG) or serine deaminase (SDS) (24 hr). The results (k) are presented as mean ± SEM (n = 3). Significance was determined by unpaired and two-tailed Student’s t-test or one-way ANOVA with Dunnett’s multiple comparisons adjustment.
Extended Data Fig. 4 GLS is correlated with SREBP-1 in human tumour tissues.
a, Representative IHC images of anti-GLS and -SREBP-1 staining in tumour vs. adjacent normal tissues from individuals with adenocarcinoma (Adeno) or squamous lung cancer. Scale bars, 50 μm. b, c, Representative IHC images of anti-GLS and anti-SREBP-1 staining from lung cancer TMA (b). Representative images of different levels of anti-GLS or anti-SREBP-1 staining and scoring are shown in (c). d, Comparison of GLS expression and SREBP-1 levels in 50 paired tumours vs. adjacent normal lung tissues from the lung cancer TMA based on H score. Significance was determined by an unpaired Student’s t-test. e, Genetic inhibition of GLS or SREBP-1 dramatically suppressed lung tumour growth in vivo. NSCLC H1299 cells were infected with shGLS- or shSREBP-1-expressing lentivirus for 48 hr and then were implanted (2 ×106 cells/mouse) into the flank of nude mice. The tumours were isolated from mice at 53 days post-implantation and were imaged (left panel) and weighed (right panel) for comparison. Data are shown as mean ± SEM (n = 6). Significance was determined by one-way ANOVA with Dunnett’s multiple comparisons adjustment. f, Representative IHC images of anti-GLS, anti-SREBP-1, anti-ASPG and anti-SDS staining in tumour tissues from patients with GBM. Scale bars, 50 μm. g, h, Representative images of anti-GLS and anti-SREBP-1 staining from glioma TMA (g). Representative images of different levels of anti-GLS or anti-SREBP-1 staining and scoring are shown in (h).
Extended Data Fig. 5 Ammonia binds to SCAP stimulating SCAP/SREBP activation.
a, Representative confocal images of U87 cells in response to glutamine (4 mM), glucose (5 mM) or NH4Cl (4 mM) stimulation for 12 hr with/without CB-839 (100 nM) under serum-free culture conditions. Scale bars, 10 μm. b, In vitro SCAP ER-budding assay. H1299 cells were stimulated with/without glutamine (4 mM) or NH4Cl (4 mM) for 4 hr under serum-free medium (5 mM glucose). Microsomes were purified and incubated at 37 °C for 15 min or on ice (as time 0) with cytosol extracts from rat liver in the presence of ATP and GTP (left panel). Alternatively, microsomes purified from H1299 cells cultured with glucose (5 mM) alone (2 hr) were incubated with NH4Cl (1 mM) or NaCl (1 mM) at 37 °C or on ice together with liver extracts as above (right panel). The mixtures were centrifuged to separate budded vesicles from the ER membrane fractions, which were then analyzed by Western blot by using indicated antibodies. c, Co-solvent NH3 computational mapping of SCAP. d, Alignment of the SCAP protein fragment. e, A schematic model for the sequential binding of NH4+ to SCAP obtained from the co-solvent ammonia mapping and NH4+-bound SCAP simulations. f, Western blot analysis of HEK293T cells transfected with GFP, wild-type or different GFP-SCAP mutants together with full-length Flag-SREBP-1c for 24 hr and then stimulated with glutamine (4 mM) for 12 hr under serum-free conditions (5 mM glucose). g. Co-solvent ammonia mapping for SCAP bound with 25-HC. Right panel shows the biochemical analysis of GFP-SCAP-bound ammonia in HEK293T cells stimulated with NH4Cl (4 mM) for 2 hr with/without pretreatment with 25-HC (10 µg/ml, 1 hr) using an ammonia assay kit. Top panel shows by western blot that equal amounts of proteins were purified. The results are presented as mean ± SEM (n = 3). Significance was determined by unpaired and two-tailed Student’s t-test. h, i, Western blot analysis of H1299 cells cultured with NH4Cl (4 mM) (h) or glutamine (4 mM) (i) for 12 hr in serum-free medium (5 mM glucose) together with a cholesterol/25-hydroxycholesterol mixture (sterols).
Extended Data Fig. 6 Ammonia binding induces SCAP dissociation from Insig.
a–d, Comparison of the coupling, tilting and kink angles of S3, S5 and S6 helices during the 1 μs simulations of SCAP bound with NH4+ vs. SCAP without bound NH4+. In panel (a), S3 and S6 helices from the NH4+ unbound simulation (in light gray) is aligned with the NH4+ bound simulation (in dark gray). NH4+, D428, S326 and S330 are shown in the stick representation. The coupling of the S3 and S6 helices was altered by the binding of NH4+ (a). In the NH4+ bound simulation, the S3 helix had a smaller tilting angle (b) and S5 and S6 helix had a larger tilting angle (c and d). Inset in panel (b) illustrates a helix titling angle. Insets in panel (c) and (d) illustrate a helix kink conformation with the lower part of the helix aligned (white), and the top part of the helix showing a difference between NH4+ bound and NH4+ unbound SCAP. Only converged data from the last 500 ns of each simulation were used for the histogram analysis. e, Comparison of the interface contact area between SCAP and Insig during the simulations of the NH4+ bound SCAP vs. the NH4+ unbound SCAP. f, A schematic model for NH4+ regulated SCAP activation. Left: Insig-SCAP binding in the absence of 25-HC and NH4+. Top: Binding of 25-HC blocks NH4+ binding to prevent SCAP activation (orange). Middle: Absence of 25-HC opens the channel, which permits the entry of NH4+ to bind to D428 first, then to S326/S330 to form a stable binding site, leading to significant conformational changes of SCAP (red) and its dissociation from Insig for subsequent translocation and SREBP activation. Bottom: D428A mutant is unable to bind NH4+, preventing NH4+ from inducing conformational changes required for SCAP dissociation from Insig in the absence of 25-HC; thus, it cannot be activated by NH4+.
Extended Data Fig. 7 SCAP D428A mutation completely abolishes ammonia function.
a, Representative confocal microscopy images of wild-type or mutant (D428A) GFP-SCAP in U87 cells compared to the Golgi marker Giantin (red) in response to glutamine or NH4Cl stimulation in the presence of glucose. U87 cells were cultured on coverslips in DMEM medium supplemented with 5% FBS for 24 hr, followed by transfection with wild-type or mutant (D428A) GFP-SCAP plasmids for 24 hr. The transfected cells were washed with PBS once and incubated with glutamine (4 mM) or NH4Cl (4 mM) for 12 hr in fresh serum-free DMEM medium with the presence of glucose (5 mM). Cell culture conditions prior to treatment are the same for subsequent panels. Scale bars, 10 μm. b, Western blot analysis of membrane and nuclear extracts from HEK293T cells transfected with GFP, GFP-SCAP wild-type or D428A mutant plasmids at the indicated doses together with full-length Flag-SREBP-1c for 24 hr and then placed in fresh serum-free DMEM medium containing glutamine (4 mM) and glucose (5 mM) for another 12 hr. c, Western blot analysis of membrane and nuclear extracts from HEK293T cells transfected with GFP, GFP-SCAP wild-type or mutant D428A, D428E (glutamate), D428N (asparagine), D428K (lysine) together with full-length Flag-SREBP-1c for 24 hr and then placed in fresh serum-free medium in the presence of glucose (5 mM) and glutamine (4 mM) for another 12 hr. d, e, Western blot analysis of membrane (for GFP-SCAP and SREBP precursors) and nuclear extracts (for N-terminal SREBPs) from HEK293T cells transfected with GFP, wild-type or mutant GFP-SCAP (D428A) together with full-length Flag-SREBP-1a (d) or HA-SREBP-2 (e) for 24 hr and then stimulated with glutamine (4 mM) or NH4Cl (4 mM) in the presence of glucose (5 mM) under fresh serum-free medium.
Extended Data Fig. 8 D428A mutation abolishes SCAP-promoted tumour growth.
a, b, Gross and macroscopic images of mouse lungs (a) and H&E staining of lung sections (b) at day 50 after mouse implantation with H1299 cells expressing GFP, wild-type (WT) or mutant GFP-SCAP D428A. Framed images in red were presented in Fig. 6d as representatives. Scale bars, 2 mm. The number of nodules on mice lung sections was quantified by ImageJ (b, lower panel). Data are shown as mean ± SEM (n = 5). Significance was determined by one-way ANOVA with Dunnett’s multiple comparisons adjustment. c, MRI scans of mouse brain at day 12 after implantation of GBM30 cells stably transfected with GFP, wild-type or mutant (D428A) GFP-SCAP (3.5 × 103 cells/mouse). Yellow circles indicate tumour location. White arrows indicate injection site. Scatter plot shows tumour volume from MRI scans quantified from the outlined region-of-interest (ROIs) (right panel). The results are presented as mean ± SEM (n = 5). Significance was determined by unpaired and two-tailed Student’s t-test. d, H&E staining of mouse brain sections excised at day 17 after implantation of GBM30 cells as described in (c). Rectangle-framed images were used in Fig. 6h as representatives. Scale bars, 1 mm.
Supplementary information
Supplementary Information
Supplementary Results, Supplementary Discussion, Supplementary Methods, Supplementary Tables and Supplementary References.
Supplementary Data 1
mRNA-seq data.
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical Source Data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical Source Data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical Source Data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, C., Geng, F., Li, Z. et al. Ammonia stimulates SCAP/Insig dissociation and SREBP-1 activation to promote lipogenesis and tumour growth. Nat Metab 4, 575–588 (2022). https://doi.org/10.1038/s42255-022-00568-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-022-00568-y
This article is cited by
-
The implications of FASN in immune cell biology and related diseases
Cell Death & Disease (2024)
-
PPARγ alleviates preeclampsia development by regulating lipid metabolism and ferroptosis
Communications Biology (2024)
-
Physiological and pathological roles of lipogenesis
Nature Metabolism (2023)
-
Targeting lipid metabolism for ferroptotic cancer therapy
Apoptosis (2023)
-
Advances in regulation and function of stearoyl-CoA desaturase 1 in cancer, from bench to bed
Science China Life Sciences (2023)