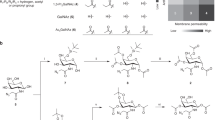
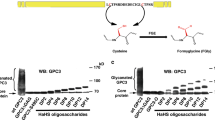
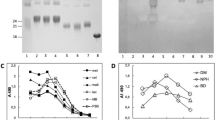
Abstract
Metabolic labeling of glycans with clickable unnatural sugars has enabled glycan analysis in multicellular systems. However, cell-type-specific labeling of glycans in vivo remains challenging. Here we develop genetically encoded metabolic glycan labeling (GeMGL), a cell-type-specific strategy based on a bump-and-hole pair of an unnatural sugar and its matching engineered enzyme. N-pentynylacetylglucosamine (GlcNAl) serves as a bumped analog of N-acetylglucosamine (GlcNAc) that is specifically incorporated into glycans of cells expressing a UDP-GlcNAc pyrophosphorylase mutant, AGX2F383G. GeMGL with the 1,3-di-O-propionylated GlcNAl (1,3-Pr2GlcNAl) and AGX2F383G pair was demonstrated in cell cocultures, and used for specific labeling of glycans in mouse xenograft tumors. By generating a transgenic mouse line with AGX2F383G expressed under a cardiomyocyte-specific promoter, we performed specific imaging of cardiomyocyte glycans in the heart and identified 582 cardiomyocyte O-GlcNAcylated proteins with no interference from other cardiac cell types. GeMGL will facilitate cell-type-specific glycan imaging and glycoproteomics in various tissues and disease models.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available within the paper and Supplementary Information. The proteomic raw data are deposited on ProteomeXchange with the dataset identifier PXD030663. Source data are provided with this paper.
References
Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Comelli, E. M. et al. A focused microarray approach to functional glycomics: transcriptional regulation of the glycome. Glycobiology 16, 117–131 (2006).
Sun, S. et al. N-GlycositeAtlas: a database resource for mass spectrometry-based human N-linked glycoprotein and glycosylation site mapping. Clin. Proteom. 16, 35 (2019).
Reily, C., Stewart, T. J., Renfrow, M. B. & Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 (2019).
Xie, R., Hong, S. & Chen, X. Cell-selective metabolic labeling of biomolecules with bioorthogonal functionalities. Curr. Opin. Chem. Biol. 17, 747–752 (2013).
Alvarez-Castelao, B. et al. Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat. Biotechnol. 35, 1196–1201 (2017).
Krogager, T. P. et al. Labeling and identifying cell-specific proteomes in the mouse brain. Nat. Biotechnol. 36, 156–159 (2018).
Prescher, J. A., Dube, D. H. & Bertozzi, C. R. Chemical remodelling of cell surfaces in living animals. Nature 430, 873–877 (2004).
Chang, P. V. et al. Copper-free click chemistry in living animals. Proc. Natl Acad. Sci. USA 107, 1821–1826 (2010).
Rong, J. et al. Glycan imaging in intact rat hearts and glycoproteomic analysis reveal the upregulation of sialylation during cardiac hypertrophy. J. Am. Chem. Soc. 136, 17468–17476 (2014).
Xie, R., Hong, S., Feng, L., Rong, J. & Chen, X. Cell-selective metabolic glycan labeling based on ligand-targeted liposomes. J. Am. Chem. Soc. 134, 9914–9917 (2012).
Xie, R. et al. Targeted imaging and proteomic analysis of tumor‐associated glycans in living animals. Angew. Chem. Int. Ed. 53, 14082–14086 (2014).
Sun, Y. et al. Mechanistic investigation and multiplexing of liposome-assisted metabolic glycan labeling. J. Am. Chem. Soc. 140, 3592–3602 (2018).
Chang, P. V., Dube, D. H., Sletten, E. M. & Bertozzi, C. R. A strategy for the selective imaging of glycans using caged metabolic precursors. J. Am. Chem. Soc. 132, 9516–9518 (2010).
Shim, M. K. et al. Cathepsin B‐specific metabolic precursor for in vivo tumor‐specific fluorescence imaging. Angew. Chem. Int. Ed. 55, 14698–14703 (2016).
Wang, H. et al. Selective in vivo metabolic cell-labeling-mediated cancer targeting. Nat. Chem. Biol. 13, 415–424 (2017).
Woo, C. M., Iavarone, A. T., Spiciarich, D. R., Palaniappan, K. K. & Bertozzi, C. R. Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 12, 561–567 (2015).
Qin, K. et al. Quantitative profiling of protein O-GlcNAcylation sites by an isotope-tagged cleavable linker. ACS Chem. Biol. 13, 1983–1989 (2018).
Boyce, M. et al. Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl Acad. Sci. 108, 3141–3146 (2011).
Yu, S.-H. et al. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc. Natl Acad. Sci. USA 109, 4834–4839 (2012).
Schumann, B. et al. Bump-and-hole engineering identifies specific substrates of glycosyltransferases in living cells. Mol. Cell 78, 824–834.e15 (2020).
Qin, W. et al. Artificial cysteine S‐glycosylation induced by per‐O‐acetylated unnatural monosaccharides during metabolic glycan labeling. Angew. Chem. Int. Ed. 57, 1817–1820 (2018).
Qin, K., Zhang, H., Zhao, Z. & Chen, X. Protein S-glyco-modification through an elimination–addition mechanism. J. Am. Chem. Soc. 142, 9382–9388 (2020).
Hao, Y. et al. Next-generation unnatural monosaccharides reveal that ESRRB O-GlcNAcylation regulates pluripotency of mouse embryonic stem cells. Nat. Commun. 10, 4065 (2019).
Zaro, B. W., Yang, Y.-Y., Hang, H. C. & Pratt, M. R. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc. Natl Acad. Sci. USA 108, 8146–8151 (2011).
Batt, A. R., Zaro, B. W., Navarro, M. X. & Pratt, M. R. Metabolic chemical reporters of glycans exhibit cell‐type‐selective metabolism and glycoprotein labeling. Chem. Bio. Chem. 18, 1177–1182 (2017).
Cioce, A. et al. Optimization of metabolic oligosaccharide engineering with Ac4GalNAlk and Ac4GlcNAlk by an engineered pyrophosphorylase. ACS Chem. Biol. 16, 1961–1967 (2021).
Subramaniam, A. et al. Tissue-specific regulation of the α-myosin heavy chain gene promoter in transgenic mice. J. Biol. Chem. 266, 24613–24620 (1991).
Dassanayaka, S. & Jones, S. P. O-GlcNAc and the cardiovascular system. Pharm. Therapeut. 142, 62–71 (2014).
Wright, J. N., Collins, H. E., Wende, A. R. & Chatham, J. C. O-GlcNAcylation and cardiovascular disease. Biochem Soc. T 45, 545–553 (2017).
Zielinska, D. F., Gnad, F., Wiśniewski, J. R. & Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 (2010).
Liu, M.-Q. et al. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 8, 438 (2017).
Alfaro, J. F. et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl Acad. Sci. USA 109, 7280–7285 (2012).
Trinidad, J. C. et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell Proteom. 11, 215–229 (2012).
Aguilar, A. L. et al. Profiling of protein O-GlcNAcylation in murine CD8+ effector- and memory-like T cells. ACS Chem. Biol. 12, 3031–3038 (2017).
Dube, D. H., Prescher, J. A., Quang, C. N. & Bertozzi, C. R. Probing mucin-type O-linked glycosylation in living animals. Proc. Natl Acad. Sci. USA 103, 4819–4824 (2006).
Xie, R. et al. In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy. Proc. Natl Acad. Sci. USA 113, 5173–5178 (2016).
Wang-Gillam, A., Pastuszak, I. & Elbein, A. D. A 17-amino acid insert changes UDP-N-acetylhexosamine pyrophosphorylase specificity from UDP-GalNAc to UDP-GlcNAc. J. Biol. Chem. 273, 27055–27057 (1998).
Peneff, C. et al. Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J. 20, 6191–6202 (2001).
Chen, Y. et al. Quantitative profiling of protein carbonylations in ferroptosis by an aniline-derived probe. J. Am. Chem. Soc. 140, 4712–4720 (2018).
Wen, L. et al. Chemoenzymatic synthesis of unnatural nucleotide sugars for enzymatic bioorthogonal labeling. ACS Catal. 8, 7659–7666 (2018).
Nakajima, K. et al. Simultaneous determination of nucleotide sugars with ion-pair reversed-phase HPLC. Glycobiology 20, 865–871 (2010).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
This project is supported by the National Key Research and Development Program of China (grant no. 2018YFA0507600) and the National Natural Science Foundation of China (grant nos. 22037001, 9215330 and 91753206).
Author information
Authors and Affiliations
Contributions
X.F., Q.S. and X.C. conceived the project. X.F. and Q.S. prepared all samples and performed biochemistry, cell biology and animal experiments. D.S., Q.S. and X.F. performed fluorescence imaging experiments. Y.H. and X.F. generated stable cell lines. X.F., Q.S. and J.W. performed chemoproteomics experiments. X.F. and C.W. synthesized organic compounds. X.F., Q.S. and X.C. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Wesley Zandberg and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Fluorescence imaging of MCF7 cells metabolically labeled with GlcNAz and 1,3-Pr2GlcNAz.
a-i, MCF7-T2A-GFP cells (a and b), MCF7-AGX2-T2A-GFP cells (c and d), MCF7-AGX2F381G-T2A-GFP cells (f and g), MCF7–AGX2F383G-T2A-GFP cells (h and i), or MCF7 cells transiently expressing AGX1-GFP or AGX2-GFP (e) were incubated with vehicle, 1 mM GlcNAz, or 100 μM 1,3-Pr2GlcNAz for 48 h. For imaging of cell-surface glycans (a, c, f, and h), the cells were directly reacted with alkyne-TAMRA, followed by imaging by confocal fluorescence microscopy. For imaging of intracellular glycans (b, d, e, g and i), the cells were fixed, permeabilized, reacted with alkyne-TAMRA, and visualized using confocal fluorescence microscopy. The nuclei were stained with Hoechst 33342 (blue). In a, the fluorescence intensity of TAMRA in the boxed areas was adjusted to 3 times, so that the weak cell-surface labeling can be observed. Scale bars, 20 μm. Representative results are shown from three independent experiments.
Extended Data Fig. 2 In-gel fluorescence scanning analysis of lysates from cells incubated with GlcNAz or 1,3-Pr2GlcNAz.
a-d, MCF7-T2A-GFP (a), AGX2-T2A-GFP (b), AGX2F381G-T2A-GFP (c), and AGX2F383G-T2A-GFP cells (d) were treated with vehicle, GlcNAz or 1,3-Pr2GlcNAz at varied concentrations for 48 h, followed by reacted with alkyne-Cy5 and analysis by in-gel fluorescence scanning. Coomassie Brilliant Blue (CBB)-stained gels demonstrate equal loading. Western blots demonstrate comparable expression of AGX2F383G by using an antibody against the N-terminal Flag tag. Representative results are shown from three independent experiments.
Extended Data Fig. 3 Fluorescence imaging of MCF7 cells metabolically labeled with GlcNAl and 1,3-Pr2GlcNAl.
a-f, MCF7-T2A-GFP (a), MCF7-AGX2-T2A-GFP (b and c), MCF7-AGX2F383G-T2A-GFP (d), or MCF7-AGX2F381G-T2A-GFP cells (e and f) were incubated with vehicle, 1 mM GlcNAl, or 100 μM 1,3-Pr2GlcNAl for 48 h. For imaging of cell-surface glycans (a, b, d, and e), the cells were directly reacted with azide-TAMRA, followed by imaging by confocal fluorescence microscopy. For imaging of intracellular glycans (c and f), the cells were fixed, permeabilized, reacted with azide-TAMRA, and visualized by confocal fluorescence microscopy. The nuclei were stained with Hoechst 33342 (blue). Scale bars, 20 μm. Representative results are shown from three independent experiments.
Extended Data Fig. 4 In-gel fluorescence scanning analysis of lysates from cells incubated with GlcNAl or 1,3-Pr2GlcNAl.
MCF7-AGX2-T2A-GFP (a) and MCF7-AGX2F381G-T2A-GFP (b) cells were treated with vehicle, GlcNAl or 1,3-Pr2GlcNAl at varied concentrations for 48 h, followed by reacted with azide-Cy5 and analysis by in-gel fluorescence scanning. Coomassie Brilliant Blue (CBB)-stained gels demonstrate equal loading. Western blots demonstrate comparable expression of AGX2 or AGX2F381G by using an antibody against the N-terminal Flag tag. Representative results are shown from three independent experiments.
Extended Data Fig. 5 Enzymatic conversion of GlcNAl-1-P to UDP-GlcNAl in vitro.
a, TLC analysis of conversion from GlcNAl-1-P to UDP-GlcNAl. GlcNAl-1-P at 10 mM was incubated with 10 μg/mL AGX2 (2), AGX2F381G (3), or AGX2F383G (4) at 37 °C for 30 min, followed by heating at 90 °C for 2 min to quench the reaction. The TLC plates were developed using the solvent (n-butanol:acetic acid:H2O = 2:1:1), and stained with p-anisaldehyde. The standard nucleotide sugars GlcNAl-1-P and UDP-GlcNAl were shown in line 1 and 5, respectively. b, Mass spectra showing MW of the enzymatic production matching UDP-GlcNAl. c, Kinetic analysis of different enzymes in catalyzing formation of UDP-GlcNAl. AGX2, AGX2F381G, or AGX2F383G was incubated with GlcNAl-1-P at varied concentrations for 30 min at 37 °C, quenched, and analyzed by HPLC. Error bars represent mean ± s.d.. Results are from three independent experiments.
Extended Data Fig. 6 Metabolism of 1,3-Pr2GlcNAl in cells.
a, HPAEC traces of extracts from MCF7-AGX2F383G-T2A-GFP cells fed with vehicle or 1,3-Pr2GlcNAl at varied concentrations and from MCF7-T2A-GFP cells treated with vehicle for 36 h. The nucleotides sugars were extracted by solid-phase extraction chromatography. ADP-glucose (ADP-Glc) was used as a spike-in control to normalize the UV absorbance between different runs. The inserted traces show the retention time of nucleotide sugar standards in separate runs. Representative results are shown from three independent experiments. b, Quantification of the cellular level of nucleotides. A.U., arbitrary units. Error bars represent mean ± s.d. from three independent experiments. The P values listed were calculated using the two-tailed Student's t-test. **P<0.01 (one-way ANOVA).
Extended Data Fig. 7 Fluorescence imaging of the cocultured cells metabolically labeled with GlcNAl, 1,3-Pr2GlcNAl, GlcNAz, or 1,3-Pr2GlcNAz.
a-e, MCF7 cells stably expressing AGX2-T2A-GFP (a and c), AGX2F381G-T2A-GFP (b and d), and AGX2F383G-T2A-GFP (e) were cocultured with HeLa cells, respectively. The cocultures were incubated with vehicle, 1 mM GlcNAl, or 100 μM 1,3-Pr2GlcNAl (a and b); the cocultures were incubated with vehicle, 1 mM GlcNAz, or 100 μM 1,3-Pr2GlcNAz (c, d, and e). After incubation for 48 h, the cells were fixed, permeabilized, and reacted with azide-TAMRA or alkyne-TAMRA. The nuclei were stained with Hoechst 33342 (blue). Scale bars, 20 μm. Representative results are shown from three independent experiments.
Extended Data Fig. 8 Confocal fluorescence images of the cardiac tissue slices from the α-MHC-AGX2F383G mice administered with 1,3-Pr2GlcNAl, related to Fig. 4c.
The slices were click-labeled with azide-TAMRA, immunostained with an anti-cTnT antibody, and stained with Hoechst 33343. Magnified views of three boxed regions are shown in the three rows below, respectively. Scale bars, 20 μm. Representative results are shown from three independent experiments.
Extended Data Fig. 9 Evaluation of the effect of AGX2F383G expression on protein O-GlcNAcylation.
a,b, Immunoblots showing O-GlcNAcylated proteins of MCF7-T2A-GFP and MCF7-AGX2F383G-T2A-GFP cells (a), or from the heart tissue of WT and α-MHC-AGX2F383G mice (b). CTD110.6 is an O-GlcNAc-specific antibody. Immunoblots using an anti-Flag antibody show the expression of AGX2F383G. CBB-stained gels are shown as the loading control. Representative results are shown from three independent experiments.
Extended Data Fig. 10 Cardiac glycoproteins identified from α-MHC-AGX2F383G mice injected with 1,3-Pr2GlcNAl.
a,b,c Volcano plots displaying the average log2 enrichment ratios for proteins quantified in at least two out of three biological replicates (x axis) and their adjusted P values (y axis; empirical Bayes moderated t test, adjusted by the Benjamini-Hochberg method). Proteins with adjusted P value < 0.01 and a fold change of two or greater were considered as identified glycoproteins. 967 and 949 proteins were identified over the vehicle (heavy/light, H/L; a) and non-transgenic mice controls (heavy/medium, H/M; b), respectively. No protein was identified in non-transgenic mice (medium/light, M/L; c).
Supplementary information
Supplementary Information
Supplementary Figs. 1–10, Note 1 and Protocols.
Supplementary Table 1
Cardiac glycoproteins identified from α-MHC-AGX2F383G mice.
Supplementary Table 2
Identified O-GlcNAc sites in MCF7-AGX2F383G-T2A-GFP cells with 1,3-Pr2GlcNAl.
Supplementary Table 3
Identified S-glyco-modification sites in MCF7-AGX2F383G-T2A-GFP cells with 1,3-Pr2GlcNAl.
Supplementary Data 1
Statistical source data for Supplementary Fig. 5.
Source data
Source Data Fig. 1
Unprocessed western blots and gels of Fig. 1.
Source Data Fig. 3
Unprocessed western blots and gels of Fig. 3.
Source Data Fig. 4
Unprocessed western blots and gels of Fig. 4.
Source Data Extended Data Fig. 2
Unprocessed western blots and gels of Extended Data Fig. 2.
Source Data Extended Data Fig. 4
Unprocessed western blots and gels of Extended Data Fig. 4.
Source Data Extended Data Fig. 6
Statistical source data for Extended Data Fig. 6.
Source Data Extended Data Fig. 9
Unprocessed western blots and gels of Extended Data Fig. 9.
Rights and permissions
About this article
Cite this article
Fan, X., Song, Q., Sun, De. et al. Cell-type-specific labeling and profiling of glycans in living mice. Nat Chem Biol 18, 625–633 (2022). https://doi.org/10.1038/s41589-022-01016-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01016-4
This article is cited by
-
Genetically targeted chemical assembly
Nature Reviews Bioengineering (2023)
-
Cell-specific bioorthogonal tagging of glycoproteins
Nature Communications (2022)