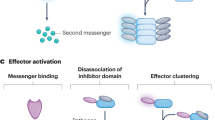
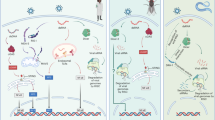
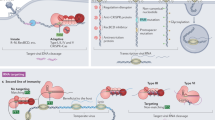
Abstract
The cell-autonomous innate immune system enables animal cells to resist viral infection. This system comprises an array of sensors that, after detecting viral molecules, activate the expression of antiviral proteins and the interferon response. The repertoire of immune sensors and antiviral proteins has long been considered to be derived from extensive evolutionary innovation in vertebrates, but new data challenge this dogma. Recent studies show that central components of the cell-autonomous innate immune system have ancient evolutionary roots in prokaryotic genes that protect bacteria from phages. These include the cyclic GMP–AMP synthase (cGAS)–stimulator of interferon genes (STING) pathway, Toll/IL-1 receptor (TIR) domain-containing pathogen receptors, the viperin family of antiviral proteins, SAMHD1-like nucleotide-depletion enzymes, gasdermin proteins and key components of the RNA interference pathway. This Perspective details current knowledge of the elements of antiviral immunity that are conserved from bacteria to humans, and presents possible evolutionary scenarios to explain the observed conservation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Randow, F., MacMicking, J. D. & James, L. C. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science 340, 701–706 (2013).
Schoggins, J. W. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 6, 567–584 (2019).
Litman, G. W., Cannon, J. P. & Dishaw, L. J. Reconstructing immune phylogeny: new perspectives. Nat. Rev. Immunol. 5, 866–879 (2005).
Cohen, D. et al. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574, 691–695 (2019).
Bernheim, A. et al. Prokaryotic viperins produce diverse antiviral molecules. Nature 589, 120–124 (2021).
Johnson, A. G. et al. Bacterial gasdermins reveal an ancient mechanism of cell death. Science 375, 221–225 (2022).
Ofir, G. et al. Antiviral activity of bacterial TIR domains via immune signaling molecules. Nature 600, 116–120 (2021).
Swarts, D. C. et al. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 21, 743–753 (2014).
Tal, N. et al. Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat. Microbiol. (in the press).
Bernheim, A. & Sorek, R. The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Microbiol. Rev. 4, 2744–2747 (2019).
Doron, S. et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120 (2018).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Keating, S. E., Baran, M. & Bowie, A. G. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 32, 574–581 (2011).
Hornung, V. & Latz, E. Intracellular DNA recognition. Nat. Rev. Immunol. 10, 123–130 (2010).
Ablasser, A. et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013).
Gao, P. et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP–AMP synthase. Cell 153, 1094–1107 (2013).
Diner, E. J. et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 3, 1355–1361 (2013).
Wu, J. et al. Cyclic GMP–AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–831 (2013).
Ablasser, A. & Chen, Z. J. cGAS in action: expanding roles in immunity and inflammation. Science 363, eaat8657 (2019).
Hopfner, K. P. & Hornung, V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 (2020).
Tan, X., Sun, L., Chen, J. & Chen, Z. J. Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 72, 447–478 (2018).
Li, X. D. et al. Pivotal roles of cGAS–cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 (2013).
Motwani, M., Pesiridis, S. & Fitzgerald, K. A. DNA sensing by the cGAS–STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019).
Margolis, S. R., Wilson, S. C. & Vance, R. E. Evolutionary origins of cGAS–STING signaling. Trends Immunol. 38, 733–743 (2017).
Wu, X. et al. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res. 42, 8243–8257 (2014).
Morehouse, B. R. et al. STING cyclic dinucleotide sensing originated in bacteria. Nature 586, 429–433 (2020).
Kranzusch, P. J. et al. Ancient origin of cGAS–STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903 (2015).
Davies, B. W., Bogard, R. W., Young, T. S. & Mekalanos, J. J. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370 (2012).
Krasteva, P. V. & Sondermann, H. Versatile modes of cellular regulation via cyclic dinucleotides. Nat. Chem. Biol. 13, 350–359 (2017).
Kranzusch, P. J. et al. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell 158, 1011–1021 (2014).
Whiteley, A. T. et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199 (2019).
Zhu, D. et al. Structural biochemistry of a Vibrio cholerae dinucleotide cyclase reveals cyclase activity regulation by folates. Mol. Cell 55, 931–937 (2014).
Severin, G. B. et al. Direct activation of a phospholipase by cyclic GMP–AMP in El Tor Vibrio cholerae. Proc. Natl Acad. Sci. USA 115, E6048–E6055 (2018).
Millman, A., Melamed, S., Amitai, G. & Sorek, R. Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol. 5, 1608–1615 (2020).
Lowey, B. et al. CBASS immunity uses CARF-related effectors to sense 3′–5′- and 2′–5′-sinked cyclic oligonucleotide signals and protect bacteria from phage infection. Cell 182, 38–49 (2020).
Ye, Q. et al. HORMA domain proteins and a Trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Mol. Cell 77, 709–722 (2020).
Lau, R. K. et al. Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol. Cell 77, 723–733 (2020).
Govande, A. A., Duncan-Lowey, B., Eaglesham, J. B., Whiteley, A. T. & Kranzusch, P. J. Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense. Cell Rep. 35, 109206 (2021).
Kranzusch, P. J. cGAS and CD-NTase enzymes: structure, mechanism, and evolution. Curr. Opin. Struct. Biol. 59, 178–187 (2019).
Duncan-Lowey, B., McNamara-Bordewick, N. K., Tal, N., Sorek, R. & Kranzusch, P. J. Effector-mediated membrane disruption controls cell death in CBASS antiphage defense. Mol. Cell 81, 1–13 (2021).
Burroughs, A. M., Zhang, D., Schäffer, D. E., Iyer, L. M. & Aravind, L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, 10633–10654 (2015).
Helbig, K. J. & Beard, M. R. The role of viperin in the innate antiviral response. J. Mol. Biol. 426, 1210–1219 (2014).
Seo, J. Y., Yaneva, R. & Cresswell, P. Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 10, 534–539 (2011).
Chin, K. C. & Cresswell, P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl Acad. Sci. USA 98, 15125–15130 (2001).
Rivera-Serrano, E. E. et al. Viperin reveals its true function. Annu. Rev. Virol. 7, 421–446 (2020).
Gizzi, A. S. et al. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558, 610–614 (2018).
Seifert, M. et al. Inhibition of SARS-CoV-2 polymerase by nucleotide analogs from a single-molecule perspective. eLife 10, e70968 (2021).
Fenwick, M. K., Li, Y., Cresswell, P., Modis, Y. & Ealick, S. E. Structural studies of viperin, an antiviral radical SAM enzyme. Proc. Natl Acad. Sci. USA 114, 6806–6811 (2017).
Makarova, K. S., Wolf, Y. I., Snir, S. & Koonin, E. V. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 193, 6039–6056 (2011).
Lachowicz, J. C., Gizzi, A. S., Almo, S. C. & Grove, T. L. Structural insight into the substrate scope of viperin and viperin-like enzymes from three domains of life. Biochemistry 60, 2116–2129 (2021).
Hollenbaugh, J. A. et al. Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 9, e1003481 (2013).
Baldauf, H. M. et al. SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 (2012).
Hrecka, K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011).
Laguette, N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011).
Li, N., Zhang, W. & Cao, X. Identification of human homologue of mouse IFN-γ induced protein from human dendritic cells. Immunol. Lett. 74, 221–224 (2000).
Ayinde, D., Casartelli, N. & Schwartz, O. Restricting HIV the SAMHD1 way: through nucleotide starvation. Nat. Rev. Microbiol. 10, 675–680 (2012).
Goldstone, D. C. et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 (2011).
Kondo, N. et al. Insights into different dependence of dNTP triphosphohydrolase on metal ion species from intracellular ion concentrations in Thermus thermophilus. Extremophiles 12, 217–223 (2008).
Mega, R., Kondo, N., Nakagawa, N., Kuramitsu, S. & Masui, R. Two dNTP triphosphohydrolases from Pseudomonas aeruginosa possess diverse substrate specificities. FEBS J. 276, 3211–3221 (2009).
Singh, D. et al. Structure of Escherichia coli dGTP triphosphohydrolase: a hexameric enzyme with DNA effector molecules. J. Biol. Chem. 290, 10418–10429 (2015).
Barnes, C. O. et al. The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity. Proc. Natl Acad. Sci. USA 116, 9333–9339 (2019).
Severin, G. et al. A broadly conserved deoxycytidine deaminase protects bacteria from phage infection. Preprint at bioRxiv https://doi.org/10.1101/2021.03.31.437871 (2021).
Liu, X. et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016).
Shi, J. et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015).
Kayagaki, N. et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015).
He, W. T. et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25, 1285–1298 (2015).
Bergsbaken, T., Fink, S. L. & Cookson, B. T. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7, 99–109 (2009).
Ding, J. et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 (2016).
Liu, Z. et al. Crystal structures of the full-length murine and human Gasdermin D reveal mechanisms of autoinhibition, lipid binding, and oligomerization. Immunity 51, 43–49 (2019).
Ruan, J., Xia, S., Liu, X., Lieberman, J. & Wu, H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67 (2018).
Xia, S. et al. Gasdermin D pore structure reveals preferential release of mature interleukin-1. Nature 593, 607–611 (2021).
Jiang, S., Zhou, Z., Sun, Y., Zhang, T. & Sun, L. Coral gasdermin triggers pyroptosis. Sci. Immunol. 5, eabd2591 (2020).
Daskalov, A., Mitchell, P. S., Sandstrom, A., Vance, R. E. & Glass, N. L. Molecular characterization of a fungal gasdermin-like protein. Proc. Natl Acad. Sci. USA 117, 18600–18607 (2020).
Baulcombe, D. RNAi in plants. Nature 431, 356–363 (2004).
Haasnoot, J., Westerhout, E. M. & Berkhout, B. RNA interference against viruses: strike and counterstrike. Nat. Biotechnol. 25, 1435–1443 (2007).
Ding, S. W. RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644 (2010).
Guo, Z., Li, Y. & Ding, S. W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 19, 31–44 (2019).
Wilson, R. C. & Doudna, J. A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 42, 217–239 (2013).
Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001).
Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R. & Hannon, G. J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150 (2001).
Raja, P., Jackel, J. N., Li, S., Heard, I. M. & Bisaro, D. M. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 88, 2611–2622 (2014).
Raja, P., Sanville, B. C., Buchmann, R. C. & Bisaro, D. M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 82, 8997–9007 (2008).
Li, H., Li, W. X. & Ding, S. W. Induction and suppression of RNA silencing by an animal virus. Science 296, 1319–1321 (2002).
Berkhout, B. RNAi-mediated antiviral immunity in mammals. Curr. Opin. Virol. 32, 9–14 (2018).
Ding, S.-W., Han, Q., Wang, J. & Li, W.-X. Antiviral RNA interference in mammals. Curr. Opin. Immunol. 54, 109–114 (2018).
Poirier, E. Z. et al. An isoform of Dicer protects mammalian stem cells against multiple RNA viruses. Science 373, 231–236 (2021).
Sledz, C. A., Holko, M., De Veer, M. J., Silverman, R. H. & Williams, B. R. G. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5, 834–839 (2003).
Song, J. J., Smith, S. K., Hannon, G. J. & Joshua-Tor, L. Crystal structure of argonaute and its implications for RISC slicer activity. Science 305, 1434–1437 (2004).
Parker, J. S., Roe, S. M. & Barford, D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 23, 4727–4737 (2004).
Yuan, Y. R. et al. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 19, 405–419 (2005).
Makarova, K. S., Wolf, Y. I., van der Oost, J. & Koonin, E. V. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 4, 29 (2009).
Swarts, D. C. et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261 (2014).
Olovnikov, I., Chan, K., Sachidanandam, R., Newman, D. K. & Aravin, A. A. Bacterial Argonaute samples the transcriptome to identify foreign DNA. Mol. Cell 51, 594–605 (2013).
Kuzmenko, A. et al. DNA targeting and interference by a bacterial Argonaute nuclease. Nature 587, 632–637 (2020).
Zander, A. et al. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2, 17034 (2017).
Swarts, D. C. et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 43, 5120–5129 (2015).
Hegge, J. W. et al. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 47, 5809–5821 (2019).
Sheng, G. et al. Structure-based cleavage mechanism of Thermus thermophilus argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl Acad. Sci. USA 111, 652–657 (2014).
Kuzmenko, A., Yudin, D., Ryazansky, S., Kulbachinskiy, A. & Aravin, A. A. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 47, 5822–5836 (2019).
Swarts, D. C. et al. Autonomous generation and loading of DNA guides by bacterial Argonaute. Mol. Cell 65, 985–998 (2017).
Kaya, E. et al. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl Acad. Sci. USA 113, 4057–4062 (2016).
Kropocheva, E., Kuzmenko, A., Aravin, A. A., Esyunina, D. & Kulbachinskiy, A. A programmable pAgo nuclease with universal guide and target specificity from the mesophilic bacterium Kurthia massiliensis. Nucleic Acids Res. 49, 4054–4065 (2021).
Garb, J. et al. Multiple phage resistance systems inhibit infection via SIR2-dependent NAD+ depletion. Preprint at bioRxiv https://doi.org/10.1101/2021.12.14.472415 (2021).
Zaremba, M. et al. SIR2-domain associated short prokaryotic Argonautes provide defence against invading mobile genetic elements through NAD+ depletion. Preprint at bioRxiv https://doi.org/10.1101/2021.12.14.472599 (2021).
Zeng, Z. et al. A short prokaryotic argonaute cooperates with membrane effector to confer antiviral defense. Preprint at bioRxiv https://doi.org/10.1101/2021.12.09.471704 (2021).
Koonin, E. V. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol. Direct 12, 5–14 (2017).
Shabalina, S. A. & Koonin, E. V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 23, 578–587 (2008).
Leulier, F. & Lemaitre, B. Toll-like receptors — taking an evolutionary approach. Nat. Rev. Genet. 9, 165–178 (2008).
Toshchakov, V. Y. & Neuwald, A. F. A survey of TIR domain sequence and structure divergence. Immunogenetics 72, 181–203 (2020).
Akira, S. & Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 (2004).
Brubaker, S. W., Bonham, K. S., Zanoni, I. & Kagan, J. C. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290 (2015).
Burch-Smith, T. M. & Dinesh-Kumar, S. P. The functions of plant TIR domains. Sci. STKE 2007, 1–5 (2007).
Wan, L. et al. TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365, 799–803 (2019).
Horsefield, S. et al. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365, 793–799 (2019).
Balint-Kurti, P. The plant hypersensitive response: concepts, control and consequences. Mol. Plant. Pathol. 20, 1163–1178 (2019).
Bayless, A. M. & Nishimura, M. T. Enzymatic functions for Toll/interleukin-1 receptor domain proteins in the plant immune system. Front. Genet. 11, 1–16 (2020).
Duxbury, Z. et al. Induced proximity of a TIR signaling domain on a plant-mammalian NLR chimera activates defense in plants. Proc. Natl Acad. Sci. USA 117, 18832–18839 (2020).
Tal, N. et al. Cyclic CMP and cyclic UMP mediate bacterial immunity against phages. Cell 184, 5728–5739 (2021).
Essuman, K. et al. The SARM1 Toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 93, 1334–1343 (2017).
Martin, W. F., Garg, S. & Zimorski, V. Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. B Biol. Sci. 370, 20140330 (2015).
Ku, C. et al. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524, 427–432 (2015).
Esser, C. et al. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21, 1643–1660 (2004).
Brueckner, J. & Martin, W. F. Bacterial genes outnumber archaeal genes in eukaryotic genomes. Genome Biol. Evol. 12, 282–292 (2020).
Broz, P., Pelegrín, P. & Shao, F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 20, 143–157 (2019).
De Schutter, E. et al. Punching holes in cellular membranes: biology and evolution of gasdermins. Trends Cell Biol. 31, 500–513 (2021).
Wu, J. & Chen, Z. J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 (2014).
Slavik, K. M. et al. cGAS-like receptors sense RNA and control 3′2′-cGAMP signalling in Drosophila. Nature 597, 109–113 (2021).
Civril, F. et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 (2013).
Kranzusch, P. J., Lee, A. S. Y., Berger, J. M. & Doudna, J. A. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 3, 1362–1368 (2013).
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Ebert, D. & Fields, P. D. Host–parasite co-evolution and its genomic signature. Nat. Rev. Genet. 21, 754–768 (2020).
Sackton, T. B. et al. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 39, 1461–1468 (2007).
Obbard, D. J., Jiggins, F. M., Bradshaw, N. J. & Little, T. J. Recent and recurrent selective sweeps of the antiviral RNAi gene Argonaute-2 in three species of Drosophila. Mol. Biol. Evol. 28, 1043–1056 (2011).
Söding, J. Protein homology detection by HMM–HMM comparison. Bioinformatics 21, 951–960 (2005).
Koyuncu, E. et al. Sirtuins are evolutionarily conserved viral restriction factors. mBio 5, e02249-14 (2014).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Goubau, D., Deddouche, S. & Reise Sousa, C. Cytosolic sensing of viruses. Immunity 38, 855–869 (2013).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Chen, Y. G. & Hur, S. Cellular origins of dsRNA, their recognition and consequences. Nat. Rev. Mol. Cell Biol. https://doi.org/10.1038/s41580-021-00430-1 (2021).
Chemudupati, M. et al. From APOBEC to ZAP: diverse mechanisms used by cellular restriction factors to inhibit virus infections. Biochim. Biophys. Acta Mol. Cell Res. 1866, 382–394 (2019).
Bailey, C. C., Zhong, G., Huang, I. C. & Farzan, M. IFITM-family proteins: the cell’s first line of antiviral defense. Annu. Rev. Virol. 1, 261–283 (2014).
Spence, J. S. et al. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 15, 259–268 (2019).
Neil, S. J. D., Zang, T. & Bieniasz, P. D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008).
Kieser, K. J. & Kagan, J. C. Multi-receptor detection of individual bacterial products by the innate immune system. Nat. Rev. Immunol. 17, 376–390 (2017).
Deretic, V., Saitoh, T. & Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13, 722–737 (2013).
Billings, E. A. et al. The adhesion GPCR BAI1 mediates macrophage ROS production and microbicidal activity against Gram-negative bacteria. Sci. Signal. 9, 1–13 (2016).
Kayagaki, N. et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 130, 1246–1249 (2013).
Brüssow, H. & Hendrix, R. W. Phage genomics: small is beautiful. Cell 108, 13–16 (2002).
Peterson, S. B., Bertolli, S. K. & Mougous, J. D. The central role of interbacterial antagonism in bacterial life. Curr. Biol. 30, R1203–R1214 (2020).
Hampton, H. G., Watson, B. N. J. & Fineran, P. C. The arms race between bacteria and their phage foes. Nature 577, 327–336 (2020).
Oliveira, P. H., Touchon, M. & Rocha, E. P. C. The interplay of restriction–modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 42, 10618–10631 (2014).
Tock, M. R. & Dryden, D. T. F. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8, 466–472 (2005).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Hille, F. et al. The biology of CRISPR–Cas: backward and forward. Cell 172, 1239–1259 (2018).
Gao, L. et al. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369, 1077–1084 (2020).
Millman, A. et al. Bacterial retrons function in anti-phage defense. Cell 183, 1551–1561 (2020).
Depardieu, F. et al. A eukaryotic-like serine/threonine kinase protects Staphylococci against phages. Cell Host Microbe 20, 471–481 (2016).
Owen, S. V. et al. Prophages encode phage-defense systems with cognate self-immunity. Cell Host Microbe 29, 1620–1633 (2021).
Goldfarb, T. et al. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183 (2015).
Lopatina, A., Tal, N. & Sorek, R. Abortive infection: bacterial suicide as an antiviral immune strategy. Annu. Rev. Virol. 7, 371–384 (2020).
Blanga-Kanfi, S., Amitsur, M., Azem, A. & Kaufmann, G. PrrC-anticodon nuclease: functional organization of a prototypical bacterial restriction RNase. Nucleic Acids Res. 34, 3209–3219 (2006).
Penner, M., Morad, I., Snyder, L. & Kaufmann, G. Phage T4-coded Stp: double-edged effector of coupled DNA and tRNA-restriction systems. J. Mol. Biol. 249, 857–868 (1995).
Acknowledgements
The authors thank P. Kranzusch, as well as the Sorek laboratory members, for fruitful discussion and comments on this manuscript. R.S. is supported, in part, by the European Research Council (grant ERC-AdG GA 101018520), Israel Science Foundation (grant ISF 296/21), the Deutsche Forschungsgemeinschaft (SPP 2330, grant 464312965), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine, the Minerva Foundation with funding from the Federal German Ministry for Education and Research, and the Knell Family Center for Microbiology. T.W. is supported by a Minerva Foundation postdoctoral fellowship and by a European Molecular Biology Organization (EMBO) postdoctoral fellowship (ALTF 946-2020).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
R.S. is a scientific cofounder and advisor of BiomX and Ecophage. T.W. declares no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks Z. Chen; L. Dishaw, who co-reviewed with O. Nataran; and J. van der Oost, who co-reviewed with R. Staals, for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wein, T., Sorek, R. Bacterial origins of human cell-autonomous innate immune mechanisms. Nat Rev Immunol 22, 629–638 (2022). https://doi.org/10.1038/s41577-022-00705-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-022-00705-4
This article is cited by
-
Inhibitors of bacterial immune systems: discovery, mechanisms and applications
Nature Reviews Genetics (2024)
-
Conservation and similarity of bacterial and eukaryotic innate immunity
Nature Reviews Microbiology (2024)
-
A break in mitochondrial endosymbiosis as a basis for inflammatory diseases
Nature (2024)
-
Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides
Nature Structural & Molecular Biology (2024)
-
DNA-targeting short Argonautes complex with effector proteins for collateral nuclease activity and bacterial population immunity
Nature Microbiology (2024)