Abstract
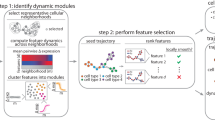
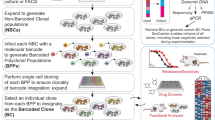
Linking single-cell genomic or transcriptomic profiles to functional cellular characteristics, in particular time-varying phenotypic changes, could help unravel molecular mechanisms driving the growth of tumour-cell subpopulations. Here we show that a custom-built optical microscope with an ultrawide field of view, fast automated image analysis and a dye activatable by visible light enables the screening and selective photolabelling of cells of interest in large heterogeneous cell populations on the basis of specific functional cellular dynamics, such as fast migration, morphological variation, small-molecule uptake or cell division. Combining such functional single-cell selection with single-cell RNA sequencing allowed us to (1) functionally annotate the transcriptomic profiles of fast-migrating and spindle-shaped MCF10A cells, of fast-migrating MDA-MB-231 cells and of patient-derived head-and-neck squamous carcinoma cells, and (2) identify critical genes and pathways driving aggressive migration and mesenchymal-like morphology in these cells. Functional single-cell selection upstream of single-cell sequencing does not depend on molecular biomarkers, allows for the enrichment of sparse subpopulations of cells, and can facilitate the identification and understanding of the molecular mechanisms underlying functional phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. Data generated in this study, including source data and the data used to make the figures, are available from Open Science Framework at https://doi.org/10.17605/OSF.IO/HG2NU. The bulk RNA-sequencing data are available at the NCBI BioProject database with the BioProject ID PRJNA751073.
Code availability
The image-analysis programme used in the study is available at https://sourceforge.net/projects/funseq.
References
Dagogo-Jack, I. & Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2017).
Bedard, P. L., Hansen, A. R., Ratain, M. J. & Siu, L. L. Tumour heterogeneity in the clinic. Nature 501, 355–364 (2013).
Lawson, D. A., Kessenbrock, K., Davis, R. T., Pervolarakis, N. & Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 20, 1349–1360 (2018).
Giladi, A. et al. Dissecting cellular crosstalk by sequencing physically interacting cells. Nat. Biotechnol. 38, 629–637 (2020).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Wan, L., Pantel, K. & Kang, Y. Tumor metastasis: moving new biological insights into the clinic. Nat. Med. 19, 1450–1464 (2013).
Chaffer, C. L. & Weinberg, R. A. A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014).
Turdo, A. et al. Meeting the challenge of targeting cancer stem cells. Front. Cell Dev. Biol. 7, 16 (2019).
Kaiser, J. The cancer stem cell gamble. Science 347, 226–229 (2015).
Lukyanov, K. A., Chudakov, D. M., Lukyanov, S. & Verkhusha, V. V. Innovation: photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol. 6, 885–891 (2005).
Zhou, X. X. & Lin, M. Z. Photoswitchable fluorescent proteins: ten years of colorful chemistry and exciting applications. Curr. Opin. Chem. Biol. 17, 682–690 (2013).
Wang, S., Moffitt, J. R., Dempsey, G. T., Xie, X. S. & Zhuang, X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc. Natl Acad. Sci. USA 111, 8452–8457 (2014).
Woll, D., Smirnova, J., Pfleiderer, W. & Steiner, U. E. Highly efficient photolabile protecting groups with intramolecular energy transfer. Angew. Chem. Int. Ed. 45, 2975–2978 (2006).
Yankaskas, C. L. et al. A microfluidic assay for the quantification of the metastatic propensity of breast cancer specimens. Nat. Biomed. Eng. 3, 452–465 (2019).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e3 (2016).
Kiselev, V. Y., Andrews, T. S. & Hemberg, M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 20, 273–282 (2019).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Puisieux, A., Brabletz, T. & Caramel, J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 16, 488–494 (2014).
Chavez, K. J., Garimella, S. V. & Lipkowitz, S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 32, 35–48 (2010).
Zhang, X. et al. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res. 19, 15 (2017).
Coradini, D., Casarsa, C. & Oriana, S. Epithelial cell polarity and tumorigenesis: new perspectives for cancer detection and treatment. Acta Pharmacol. Sin. 32, 552–564 (2011).
Amat, F. et al. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods 11, 951–958 (2014).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Chung, N. C. & Storey, J. D. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 31, 545–554 (2014).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Acknowledgements
M.-P.C. acknowledges support from the Oncode Institute, Cancer GenomiCs.nl (CGC), NWO (the Netherlands Organization for Scientific Research) Veni Grant, and Erasmus MC grant. M.-P.C. appreciates Josephine Nefkens Stichting’s support on the UFO microscope. D.B. acknowledges support by an NWO Start-up Grant (740.018.018) and ERC Starting Grant (850818 - MULTIVIsion). P.-R.S. acknowledges support from Ministry of Science and Technology (MOST) in Taiwan (Dragon Gate program: 107-2911-I-002-577 and Columbus Program: 108-2636-M-002-008- &109-2636-M-002-005-). We thank R. Agami and M. Paul for the kind gift of MCF10A-H2B-GFP and U2OS-H2B-mMaple3 cell lines, respectively; A. Theil and T. W. Kan for technical assistance with FACS sorting; J. Kraan and J. Martens for the use of their cell separation machine; the Erasmus MC Center for Biomics for the bulk-cell transcriptomic sequencing; K. T. Chen for the phototagging purification; and P. Keller for discussions about TGMM.
Author information
Authors and Affiliations
Contributions
L.Y. scripted the mTGMM cell tracking algorithm and analysed most of the image data. P.-R.S. designed and conducted experiments and performed image analysis. M.B. and M.-P.C. scripted code for analysis of the single-cell sequencing data and performed analysis. R.G.R. conducted the HNSCC experiments. R.G.R. and T.-C.C. conducted experiments on HNSCC samples provided by J.A.U.H. and R.B.d.J., and T.-C.C. performed image analysis. C.B. contributed to all the cell culture preparations for the experiments, including the gene knockdown experiments. E.v.O., F.L. and T.-C.C. contributed to the morphology detection algorithm. P.G. conducted the 3D phototagging experiment. M.M. advised on some of the single-cell experiment and sequencing analysis. D.v.S. conducted part of the cell migration and sorting experiment. D.B. and M.-P.C. set up data-acquisition hardware and software with contributions from R.v.T., S.F. and P.G. M.-P.C., D.B., L.Y., M.B. and P.-R.S. wrote the paper with input from all authors. M.-P.C. and D.B. initiated, contributed to and supervised all aspects of the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Konstantinos Konstantopoulos, Luca Magnani and Wei Li for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods, figures and references.
Rights and permissions
About this article
Cite this article
You, L., Su, PR., Betjes, M. et al. Linking the genotypes and phenotypes of cancer cells in heterogenous populations via real-time optical tagging and image analysis. Nat. Biomed. Eng 6, 667–675 (2022). https://doi.org/10.1038/s41551-022-00853-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-022-00853-x
This article is cited by
-
Bridging live-cell imaging and next-generation cancer treatment
Nature Reviews Cancer (2023)
-
Unraveling non-genetic heterogeneity in cancer with dynamical models and computational tools
Nature Computational Science (2023)