Abstract
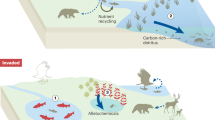
How bacterial chemotaxis is performed is much better understood than why. Traditionally, chemotaxis has been understood as a foraging strategy by which bacteria enhance their uptake of nutrients and energy, yet it has remained puzzling why certain less nutritious compounds are strong chemoattractants and vice versa. Recently, we have gained increased understanding of alternative ecological roles of chemotaxis, such as navigational guidance in colony expansion, localization of hosts or symbiotic partners and contribution to microbial diversity by the generation of spatial segregation in bacterial communities. Although bacterial chemotaxis has been observed in a wide range of environmental settings, insights into the phenomenon are mostly based on laboratory studies of model organisms. In this Review, we highlight how observing individual and collective migratory behaviour of bacteria in different settings informs the quantification of trade-offs, including between chemotaxis and growth. We argue that systematically mapping when and where bacteria are motile, in particular by transgenerational bacterial tracking in dynamic environments and in situ approaches from guts to oceans, will open the door to understanding the rich interplay between metabolism and growth and the contribution of chemotaxis to microbial life.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
01 April 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41579-022-00734-9
References
Wadhwa, N. & Berg, H. C. Bacterial motility: machinery and mechanisms. Nat. Rev. Microbiol. 20, 161–173 (2022). This recent review provides an excellent overview of the diversity in bacterial propulsion mechanisms.
Burrows, L. L. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66, 493–520 (2012).
Dufrêne, Y. F. & Persat, A. Mechanomicrobiology: how bacteria sense and respond to forces. Nat. Rev. Microbiol. 18, 227–240 (2020).
Jarrell, K. F. & McBride, M. J. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6, 466–476 (2008).
Berg, H. C. E. coli in Motion (Springer, 2004).
Bi, S. & Sourjik, V. Stimulus sensing and signal processing in bacterial chemotaxis. Curr. Opin. Microbiol. 45, 22–29 (2018).
Parkinson, J. S., Hazelbauer, G. L. & Falke, J. J. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 23, 257–266 (2015).
Porter, S. L., Wadhams, G. H. & Armitage, J. P. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9, 153–165 (2011).
Colin, R. & Sourjik, V. Emergent properties of bacterial chemotaxis pathway. Curr. Opin. Microbiol. 39, 24–33 (2017).
Brumley, D. R. et al. Cutting through the noise: bacterial chemotaxis in marine microenvironments. Front. Mar. Sci. 7, 527 (2020).
Hein, A. M., Carrara, F., Brumley, D. R., Stocker, R. & Levin, S. A. Natural search algorithms as a bridge between organisms, evolution, and ecology. Proc. Natl Acad. Sci. USA 113, 9413–9420 (2016).
Wong-Ng, J., Celani, A. & Vergassola, M. Exploring the function of bacterial chemotaxis. Curr. Opin. Microbiol. 45, 16–21 (2018).
Colin, R., Ni, B., Laganenka, L. & Sourjik, V. Multiple functions of flagellar motility and chemotaxis in bacterial physiology. FEMS Microbiol. Rev. 45, fuab038 (2021).
Schweinitzer, T. & Josenhans, C. Bacterial energy taxis: a global strategy? Arch. Microbiol. 192, 507–520 (2010).
Somavanshi, R., Ghosh, B. & Sourjik, V. Sugar influx sensing by the phosphotransferase system of Escherichia coli. PLoS Biol. 14, e2000074 (2016).
Cremer, J. et al. Chemotaxis as a navigation strategy to boost range expansion. Nature 575, 658–663 (2019). This work uses a quantitative approach to describe the classic assay of bacterial growth and migration in soft agar, and elucidates the distinct roles of attractant and nutrient in colony expansion.
Raina, J.-B., Fernandez, V., Lambert, B., Stocker, R. & Seymour, J. R. The role of microbial motility and chemotaxis in symbiosis. Nat. Rev. Microbiol. 17, 284–294 (2019). This study presents a comprehensive overview of the role of bacterial motility and chemotaxis in establishing and maintaining symbiotic relationships.
Matilla, M. A. & Krell, T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol. Rev. 42, fux052 (2018). This work presents an extensive review of the role of bacterial motility and chemotaxis in host pathogenicity from plants to animals.
Perkins, A., Tudorica, D. A., Amieva, M. R., Remington, S. J. & Guillemin, K. Helicobacter pylori senses bleach (HOCl) as a chemoattractant using a cytosolic chemoreceptor. PLoS Biol. 17, e3000395 (2019).
Tohidifar, P. et al. The unconventional cytoplasmic sensing mechanism for ethanol chemotaxis in Bacillus subtilis. mBio 11, e02177-20 (2020).
Kundu, P., Blacher, E., Elinav, E. & Pettersson, S. Our gut microbiome: the evolving inner self. Cell 171, 1481–1493 (2017).
Azam, F. & Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791 (2007).
Buchan, A., LeCleir, G. R., Gulvik, C. A. & González, J. M. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698 (2014).
Savageau, M. A. Escherichia coli habitats, cell types and molecular mechanisms of gene control. Am. Nat. 122, 732–744 (1983).
Vorholt, J. A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840 (2012).
Scharf, B. E., Hynes, M. F. & Alexandre, G. M. Chemotaxis signaling systems in model beneficial plant–bacteria associations. Plant. Mol. Biol. 90, 549–559 (2016).
Stocker, R. & Seymour, J. R. Ecology and physics of bacterial chemotaxis in the ocean. Microbiol. Mol. Biol. Rev. 76, 792–812 (2012).
Datta, M. S., Sliwerska, E., Gore, J., Polz, M. F. & Cordero, O. X. Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7, 11965 (2016).
Barbara, G. M. & Mitchell, J. G. Bacterial tracking of motile algae. FEMS Microbiol. Ecol. 44, 79–87 (2003).
Garren, M. et al. A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007 (2014).
Szurmant, H. & Ordal, G. W. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68, 301–319 (2004).
Wuichet, K. & Zhulin, I. B. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3, ra50 (2010).
Zehr, J. P., Weitz, J. S. & Joint, I. How microbes survive in the open ocean. Science 357, 646–647 (2017).
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D. & Kjelleberg, S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50 (2012).
Yawata, Y., Carrara, F., Menolascina, F. & Stocker, R. Constrained optimal foraging by marine bacterioplankton on particulate organic matter. Proc. Natl Acad. Sci. USA 117, 25571–25579 (2020). This study reveals that a marine bacterium foraging on particulate nutrient hotspots optimizes nutrient uptake using rapid switches between chemotactic and non-motile lifestyles.
Paul, K., Nieto, V., Carlquist, W. C., Blair, D. F. & Harshey, R. M. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38, 128–139 (2010).
Fenchel, T. Microbial behavior in a heterogeneous world. Science 296, 1068–1071 (2002).
Stocker, R. Marine microbes see a sea of gradients. Science 338, 628–633 (2012).
McDonald, D. E., Pethick, D. W., Mullan, B. P. & Hampson, D. J. Increasing viscosity of the intestinal contents alters small intestinal structure and intestinal growth, and stimulates proliferation of enterotoxigenic Escherichia coli in newly-weaned pigs. Br. J. Nutr. 86, 487–498 (2001).
Berg, H. C. & Turner Movement of microorganisms in viscous environments. Nature 278, 349–351 (1979).
Borer, B., Tecon, R. & Or, D. Spatial organization of bacterial populations in response to oxygen and carbon counter-gradients in pore networks. Nat. Commun. 9, 769 (2018).
Whitman, W. B., Coleman, D. C. & Wiebe, W. J. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (1998).
Raynaud, X. & Nunan, N. Spatial ecology of bacteria at the microscale in soil. PLoS ONE 9, e87217 (2014).
Lindow, S. E. & Brandl, M. T. Microbiology of the phyllosphere. Appl. Env. Microbiol. 69, 9 (2003).
Fernandez, V. I., Yawata, Y. & Stocker, R. A foraging mandala for aquatic microorganisms. ISME J. 13, 563–575 (2019).
Purcell, E. M. Life at low Reynolds number. Am. J. Phys. 45, 10 (1977).
Dusenbery, D. B. Living at Micro Scale: The Unexpected Physics of Being Small (Harvard Univ. Press, 2011).
Phillips, R. & Milo, R. Cell Biology by the Numbers (Garland Science, 2015).
Darnton, N. C., Turner, L., Rojevsky, S. & Berg, H. C. On torque and tumbling in swimming Escherichia coli. J. Bacteriol. 189, 1756–1764 (2007).
Ryu, W. S., Berry, R. M. & Berg, H. C. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403, 444–446 (2000).
Chattopadhyay, S., Moldovan, R., Yeung, C. & Wu, X. L. Swimming efficiency of bacterium Escherichia coli. Proc. Natl Acad. Sci. USA 103, 13712–13717 (2006).
Sowa, Y., Hotta, H., Homma, M. & Ishijima, A. Torque–speed relationship of the Na+-driven flagellar motor of Vibrio alginolyticus. J. Mol. Biol. 327, 1043–1051 (2003).
Taylor, J. R. & Stocker, R. Trade-offs of chemotactic foraging in turbulent water. Science 338, 675–679 (2012).
Govern, C. C. & ten Wolde, P. R. Optimal resource allocation in cellular sensing systems. Proc. Natl Acad. Sci. USA 111, 17486–17491 (2014).
Sourjik, V. & Berg, H. C. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA 99, 12669–12674 (2002).
Lan, G., Sartori, P., Neumann, S., Sourjik, V. & Tu, Y. The energy–speed–accuracy trade-off in sensory adaptation. Nat. Phys. 8, 422–428 (2012).
Stouthamer, A. H. & Bettenhaussen, C. W. A continuous culture study of an ATPase-negative mutant of Escherichia coli. Arch. Microbiol. 113, 185–189 (1977).
Macnab, R. M. in Escherichia coli and Salmonella Typhimurium: Cellular and Molecular Biology Vol. 1 (eds Nerdhardt, F. et al.) 732–759 (American Society for Microbiology, 1987).
Kempes, C. P. et al. Drivers of bacterial maintenance and minimal energy requirements. Front. Microbiol. 8, 31 (2017).
Lynch, M. & Marinov, G. K. The bioenergetic costs of a gene. Proc. Natl Acad. Sci. USA 112, 15690–15695 (2015).
Hoehler, T. M. & Jørgensen, B. B. Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11, 83–94 (2013).
Boehm, A. et al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141, 107–116 (2010).
Fang, X. & Gomelsky, M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility: c-di-GMP-dependent flagellum rotation bias. Mol. Microbiol. 76, 1295–1305 (2010).
Sathyamoorthy, R. et al. To hunt or to rest: prey depletion induces a novel starvation survival strategy in bacterial predators. ISME J. 15, 109–123 (2020).
Adler, J. & Templeton, B. The effect of environmental conditions on the motility of Escherichia coli. J. Gen. Microbiol. 46, 175–184 (1967).
Berg, H. C. & Tedesco, P. M. Transient response to chemotactic stimuli in Escherichia coli. Proc. Natl Acad. Sci. USA 72, 3235–3239 (1975).
Mitchell, J. G. The influence of cell size on marine bacterial motility and energetics. Microb. Ecol. 22, 227–238 (1991).
Castro-Sowinski, S., Burdman, S., Matan, O. & Okon, Y. in Plastics from Bacteria Vol. 14 (ed. Chen, G. G.-Q.) 39–61 (Springer, 2010).
Walter, J. M., Greenfield, D., Bustamante, C. & Liphardt, J. Light-powering Escherichia coli with proteorhodopsin. Proc. Natl Acad. Sci. USA 104, 2408–2412 (2007).
Gude, S. et al. Bacterial coexistence driven by motility and spatial competition. Nature 578, 588–592 (2020). This work presents evidence for a trade-off between motility and growth, which supports bacterial diversity through spatial segregation.
Ni, B., Colin, R., Link, H., Endres, R. G. & Sourjik, V. Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. Proc. Natl Acad. Sci. USA 117, 595–601 (2020). This work systematically compares the cost and benefit of chemotaxis in spatially extended and well-mixed environments.
Li, M. & Hazelbauer, G. L. Cellular stoichiometry of the components of the chemotaxis signaling complex. J. Bacteriol. 186, 3687–3694 (2004).
Neumann, S., Hansen, C. H., Wingreen, N. S. & Sourjik, V. Differences in signalling by directly and indirectly binding ligands in bacterial chemotaxis. EMBO J. 29, 3484–3495 (2010).
Akashi, H. & Gojobori, T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl Acad. Sci. USA 99, 3695–3700 (2002).
Basan, M. et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104 (2015).
Ni, B. et al. Evolutionary remodeling of bacterial motility checkpoint control. Cell Rep. 18, 866–877 (2017).
Fraebel, D. T. et al. Environment determines evolutionary trajectory in a constrained phenotypic space. eLife 6, e24669 (2017).
Honda, T. et al. Coordination of gene expression with cell size enables Escherichia coli to efficiently maintain motility across conditions. Preprint at bioRxiv https://doi.org/10.1101/2021.05.12.443892 (2021).
Zampieri, M., Hörl, M., Hotz, F., Müller, N. F. & Sauer, U. Regulatory mechanisms underlying coordination of amino acid and glucose catabolism in Escherichia coli. Nat. Commun. 10, 3354 (2019).
Zhao, Z. et al. Frequent pauses in Escherichia coli flagella elongation revealed by single cell real-time fluorescence imaging. Nat. Commun. 9, 1885 (2018).
Zhuang, X. et al. Live‐cell fluorescence imaging reveals dynamic production and loss of bacterial flagella. Mol. Microbiol. 114, 279–291 (2020).
Chevance, F. F. V. & Hughes, K. T. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6, 455–465 (2008). This work presents a classic overview of the gene regulatory pathway that controls flagella assembly in Gram-negative bacteria.
Amsler, C. D., Cho, M. & Matsumura, P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J. Bacteriol. 175, 6238–6244 (1993).
Lopes, J. G. & Sourjik, V. Chemotaxis of Escherichia coli to major hormones and polyamines present in human gut. ISME J. 12, 2736–2747 (2018).
Yang, J. et al. Biphasic chemotaxis of Escherichia coli to the microbiota metabolite indole. Proc. Natl Acad. Sci. USA 117, 6114–6120 (2020).
Matz, C. & Jürgens, K. High motility reduces grazing mortality of planktonic bacteria. Appl. Environ. Microbiol. 71, 921–929 (2005).
Cummings, L. A., Wilkerson, W. D., Bergsbaken, T. & Cookson, B. T. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol. Microbiol. 61, 795–809 (2006).
Yuan, J. & Berg, H. C. Ultrasensitivity of an adaptive bacterial motor. J. Mol. Biol. 425, 1760–1764 (2013).
Lestas, I., Vinnicombe, G. & Paulsson, J. Fundamental limits on the suppression of molecular fluctuations. Nature 467, 174–178 (2010).
Frankel, N. W. et al. Adaptability of non-genetic diversity in bacterial chemotaxis. eLife 3, e03526 (2014).
Goldbeter, A. & Koshland, D. E. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl Acad. Sci. USA 78, 6840–6844 (1981).
Waite, A. J. et al. Non‐genetic diversity modulates population performance. Mol. Syst. Biol. 12, 895 (2016).
Fu, X. et al. Spatial self-organization resolves conflicts between individuality and collective migration. Nat. Commun. 9, 2177 (2018). This sophisticated microfluidic study reveals that a chemotactic population may travel as a cohesive unit despite strong phenotypic heterogeneity within the population.
Long, Z., Quaife, B., Salman, H. & Oltvai, Z. N. Cell–cell communication enhances bacterial chemotaxis toward external attractants. Sci. Rep. 7, 12855 (2017).
Laganenka, L., Colin, R. & Sourjik, V. Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat. Commun. 7, 12984 (2016). This study demonstrates that bacteria may chase self-generated gradients by producing quorum-sensing molecules.
Park, S. et al. Influence of topology on bacterial social interaction. Proc. Natl Acad. Sci. USA 100, 13910–13915 (2003).
Phan, T. V. et al. Bacterial route finding and collective escape in mazes and fractals. Phys. Rev. X 10, 031017 (2020).
Waite, A. J., Frankel, N. W. & Emonet, T. Behavioral variability and phenotypic diversity in bacterial chemotaxis. Annu. Rev. Biophys. 47, 595–616 (2018). This work presents a review of the mechanisms underlying behavioural variation in bacterial chemotaxis and the consequences for chemotactic performance.
Ackermann, M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 13, 497–508 (2015).
Bódi, Z. et al. Phenotypic heterogeneity promotes adaptive evolution. PLoS Biol. 15, e2000644 (2017).
Seymour, J. R., Amin, S. A., Raina, J.-B. & Stocker, R. Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2, 17065 (2017).
Weber, L., Gonzalez‐Díaz, P., Armenteros, M. & Apprill, A. The coral ecosphere: a unique coral reef habitat that fosters coral–microbial interactions. Limnol. Oceanogr. 64, 2373–2388 (2019).
Salek, M. M., Carrara, F., Fernandez, V., Guasto, J. S. & Stocker, R. Bacterial chemotaxis in a microfluidic T-maze reveals strong phenotypic heterogeneity in chemotactic sensitivity. Nat. Commun. 10, 1877 (2019).
Ford, R. M. & Lauffenburger, D. A. Measurement of bacterial random motility and chemotaxis coefficients: II. Application of single-cell-based mathematical model. Biotechnol. Bioeng. 37, 661–672 (1991).
Lambert, B. S., Fernandez, V. I. & Stocker, R. Motility drives bacterial encounter with particles responsible for carbon export throughout the ocean. Limnol. Oceanogr. Lett. 4, 113–118 (2019).
Słomka, J., Alcolombri, U., Secchi, E., Stocker, R. & Fernandez, V. I. Encounter rates between bacteria and small sinking particles. N. J. Phys. 22, 043016 (2020).
Hein, A. M. & Martin, B. T. Information limitation and the dynamics of coupled ecological systems. Nat. Ecol. Evol. 4, 82–90 (2020).
Kiorboe, T., Grossart, H.-P., Ploug, H. & Tang, K. Mechanisms and rates of bacterial colonization of sinking aggregates. Appl. Environ. Microbiol. 68, 3996–4006 (2002).
Viswanathan, G. M. et al. Optimizing the success of random searches. Nature 401, 911–914 (1999).
Korobkova, E., Emonet, T., Vilar, J. M. G., Shimizu, T. S. & Cluzel, P. From molecular noise to behavioural variability in a single bacterium. Nature 428, 574–578 (2004).
Tu, Y. & Grinstein, G. How white noise generates power-law switching in bacterial flagellar motors. Phys. Rev. Lett. 4, 208101 (2005).
Huo, H., He, R., Zhang, R. & Yuan, J. Swimming Escherichia coli explore the environment by Lévy walk. Appl. Environ. Microbiol. 87, e02429–20 (2021).
Keegstra, J. M. et al. Phenotypic diversity and temporal variability in a bacterial signaling network revealed by single-cell FRET. eLife 6, e27455 (2017).
Colin, R. & Sourjik, V. Multiple sources of slow activity fluctuations in a bacterial chemosensory network. eLife 6, e26796 (2017).
Karin, O. & Alon, U. Temporal fluctuations in chemotaxis gain implements a simulated tempering strategy for efficient navigation in complex environments. SSRN Electron. J. 24, 102796 (2021).
Carey, J. N. et al. Regulated stochasticity in a bacterial signaling network permits tolerance to a rapid environmental change. Cell 173, 196–207.e14 (2018).
Kamino, K., Keegstra, J. M., Long, J., Emonet, T. & Shimizu, T. S. Adaptive tuning of cell sensory diversity without changes in gene expression. Sci. Adv. 6, eabc1087 (2020). This study shows that a bacterial population increases chemotactic bed-hedging when environmental signals are unavailable, but suppresses the sensory diversity when a traceable signal is presented.
Bassler, B. L. & Losick, R. Bacterially speaking. Cell 125, 237–246 (2006).
Mukherjee, S. & Bassler, B. L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382 (2019).
Budrene, E. O. & Berg, H. C. Dynamics of formation of symmetrical patterns by chemotactic bacteria. Nature 376, 49–53 (1995).
Ben-Jacob, E., Cohen, I. & Levine, H. Cooperative self-organization of microorganisms. Adv. Phys. 49, 395–554 (2000).
Adler, J. Chemotaxis in bacteria. Science 153, 708–716 (1966).
Keller, E. F. & Segel, L. A. Model for chemotaxis. J. Theor. Biol. 30, 225–234 (1971).
Saragosti, J. et al. Directional persistence of chemotactic bacteria in a traveling concentration wave. Proc. Natl Acad. Sci. USA 108, 16235–16240 (2011).
Mattingly & Emonet, T. The balancing act of growth and expansion. Nature 575, 602–603 (2019).
Liu, W., Cremer, J., Li, D., Hwa, T. & Liu, C. An evolutionarily stable strategy to colonize spatially extended habitats. Nature 575, 664–668 (2019). This study reveals that chemotactic strains selected for different speeds of range expansion in semi-solid agar can stably coexist.
Hui, S. et al. Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Mol. Syst. Biol. 11, 784 (2015).
Maser, A., Peebo, K., Vilu, R. & Nahku, R. Amino acids are key substrates to Escherichia coli BW25113 for achieving high specific growth rate. Res. Microbiol. 171, 185–193 (2020).
Yang, Y. et al. Relation between chemotaxis and consumption of amino acids in bacteria. Mol. Microbiol. 96, 1272–1282 (2015). This study is a pioneering work on the relation between chemotaxis and metabolism, where the relationship between amino acid uptake preference and chemotactic affinity in E. coli and B. subtilis is studied.
Cadotte, M. W. et al. On testing the competition–colonization trade-off in a multispecies assemblage. Am. Nat. 168, 704–709 (2006).
Amarasekare, P. Competitive coexistence in spatially structured environments: a synthesis. Ecol. Lett. 6, 1109–1122 (2003).
Levins, R. & Culver, D. Regional coexistence of species and competition between rare species. Proc. Natl Acad. Sci. USA 68, 1246–1248 (1971).
Yawata, Y. et al. Competition–dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc. Natl Acad. Sci. USA 111, 5622–5627 (2014).
Narla, A. V., Cremer, J. & Hwa, T. A traveling-wave solution for bacterial chemotaxis with growth. Proc. Natl Acad. Sci. USA 118, e2105138118 (2021). This work develops a comprehensive mathematical framework describing migrating bands of bacteria driven by growth and chemotaxis that is applicable to many environments.
Bassler, B. L., Gibbons, P. J., Yu, C. & Roseman, S. Chemotaxis to chitin oligosaccharides by Vibrio furnissi. J. Biol. Chem. 266, 24268–24275 (1991).
Konishi, H., Hio, M., Kobayashi, M., Takase, R. & Hashimoto, W. Bacterial chemotaxis towards polysaccharide pectin by pectin-binding protein. Sci. Rep. 10, 3977 (2020).
Alcolombri, U. et al. Sinking enhances the degradation of organic particles by marine bacteria. Nat. Geosci. 14, 775–780 (2021).
D’Souza, G. G., Povolo, V. R., Keegstra, J. M., Stocker, R. & Ackermann, M. Nutrient complexity triggers transitions between solitary and colonial growth in bacterial populations. ISME J. 1, 1 (2021).
Nesper, J. et al. Cyclic di-GMP differentially tunes a bacterial flagellar motor through a novel class of CheY-like regulators. eLife 6, e28842 (2017).
Basan, M. et al. A universal trade-off between growth and lag in fluctuating environments. Nature 584, 470–474 (2020).
Nguyen, J. et al. A distinct growth physiology enhances bacterial growth under rapid nutrient fluctuations. Nat. Commun. 12, 3662 (2021).
Costello, E. K., Stagaman, K., Dethlefsen, L., Bohannan, B. J. M. & Relman, D. A. The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262 (2012).
Cordero, O. X. & Datta, M. S. Microbial interactions and community assembly at microscales. Curr. Opin. Microbiol. 31, 227–234 (2016).
Rusconi, R., Garren, M. & Stocker, R. Microfluidics expanding the frontiers of microbial ecology. Annu. Rev. Biophys. 43, 65–91 (2014).
Lambert, B. S. et al. A microfluidics-based in situ chemotaxis assay to study the behaviour of aquatic microbial communities. Nat. Microbiol. 2, 1344–1349 (2017).
Clerc, E. E., Raina, J.-B., Lambert, B. S., Seymour, J. & Stocker, R. In situ chemotaxis assay to examine microbial behavior in aquatic ecosystems. J. Vis. Exp. 159, e61062 (2020).
Pleška, M., Jordan, D., Frentz, Z., Xue, B. & Leibler, S. Nongenetic individuality, changeability, and inheritance in bacterial behavior. Proc. Natl Acad. Sci. USA 118, e2023322118 (2021).
Figueroa-Morales, N. et al. 3D spatial exploration by E. coli echoes motor temporal variability. Phys. Rev. X 10, 021004 (2020).
Hazelbauer, G. L. Bacterial chemotaxis: the early years of molecular studies. Annu. Rev. Microbiol. 66, 285–303 (2012).
Adler, J., Hazelbauer, G. L. & Dahl, M. M. Chemotaxis toward sugars in Escherichia coli. J. Bacteriol. 115, 824–847 (1973).
Mesibov, R. & Adler, J. Chemotaxis toward amino acids in Escherichia coli. J. Bacteriol. 112, 12 (1972).
Dekel, E. & Alon, U. Optimality and evolutionary tuning of the expression level of a protein. Nature 436, 588–592 (2005).
Erickson, D. W. et al. A global resource allocation strategy governs growth transition kinetics of Escherichia coli. Nature 551, 119–123 (2017).
Berg, H. C. & Purcell, E. M. Physics of chemoreception. Biophys. J. 20, 193–219 (1977).
Mora, T. & Wingreen, N. S. Limits of sensing temporal concentration changes by single cells. Phys. Rev. Lett. 104, 248101 (2010).
Brumley, D. R. et al. Bacteria push the limits of chemotactic precision to navigate dynamic chemical gradients. Proc. Natl Acad. Sci. USA 116, 10792–10797 (2019).
Mattingly, H. H., Kamino, K., Machta, B. B. & Emonet, T. Escherichia coli chemotaxis is information limited. Nat. Phys. 17, 1426–1431 (2021).
Clausznitzer, D., Micali, G., Neumann, S., Sourjik, V. & Endres, R. G. Predicting chemical environments of bacteria from receptor signaling. PLoS Comput. Biol. 10, e1003870 (2014).
Flores, M., Shimizu, T. S., ten Wolde, P. R. & Tostevin, F. Signaling noise enhances chemotactic drift of E. coli. Phys. Rev. Lett. 109, 148101 (2012).
Okubo, A. & Levin, S. A. Diffusion and Ecological Problems: Modern Perspectives Vol. 14 (Springer, 2001).
Fisher, R. A. The wave of advance of advantageous genes. Ann. Eugen. 7, 355–369 (1937).
Kolmogorov, A., Petrovskii, I. & Piskunov, N. Study of a diffusion equation that is related to the growth of a quality of matter and its application to a biological problem. Mosc. Univ. Math. Bull. 1, 1–26 (1937).
Giometto, A., Rinaldo, A., Carrara, F. & Altermatt, F. Emerging predictable features of replicated biological invasion fronts. Proc. Natl Acad. Sci. USA 111, 297–301 (2014).
Gandhi, S. R., Yurtsev, E. A., Korolev, K. S. & Gore, J. Range expansions transition from pulled to pushed waves as growth becomes more cooperative in an experimental microbial population. Proc. Natl Acad. Sci. USA 113, 6922–6927 (2016).
Painter, K. J. Mathematical models for chemotaxis and their applications in self-organisation phenomena. J. Theor. Biol. 481, 162–182 (2019).
Acknowledgements
The authors thank all cited authors for the work highlighted in this Review and apologize to those not cited for length reasons. They are grateful to U. Alcolombri, Z. Landry, J. Nguyen, C. Martinez-Pérez and J. Wheeler for critical reading of the manuscript, and to V. Fernandez, N. Norris and U. Sauer for stimulating discussions. They thank R. Naisbit for scientific editing. The authors were supported by the Simons Foundation through the Principles of Microbial Ecosystems (PriME) collaboration (grant 542395) and the Swiss National Science Foundation’s National Centre of Competence in Research (NCCR) Microbiomes (no. 51NF40_180575 to R.S.).
Author information
Authors and Affiliations
Contributions
J.M.K. and F.C. researched data for the article. All authors contributed to the discussion of the content, wrote the article and reviewed and edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology and the authors acknowledge Terence Hwa and the other, anonymous, reviewers for their contribution to the peer review of this work.
Additional information
We dedicate this work to the memory of Howard C. Berg (1934–2021), a true giant in illuminating the microscale world of microorganisms, who unravelled the mechanisms of bacterial sensing and locomotion to an unprecedented level. His creative insights have represented the foundation for countless chemotaxis studies over the past decades and will remain an inspiration for future discoveries in the motile lives of bacteria.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Flagella
-
Elongated, thin and stiff filaments that generate forward thrust by rotating. Multiple filaments together may form a flagellar bundle.
- Random walk
-
Movement in which steps are taken in random directions. It can be biased if the step length or orientation favours a certain direction.
- Chemoattractants
-
Chemicals that attract an organism, inducing movement towards higher concentrations of the chemical.
- Metabolism
-
The chemical reactions required to sustain living systems: breakdown of chemicals to release energy (catabolism), synthesis of biomass (anabolism) and elimination of waste chemicals.
- Chemorepellents
-
Chemicals that repel an organism, inducing movement towards lower concentrations of the chemical.
- Flagellar motor
-
A transmembrane protein complex connecting to the flagellar filaments, which converts a protonic or ionic gradient into rotary motion.
- Sensory adaptation
-
The (partial) restoration of pre-stimulus behaviour during prolonged stimulation.
- Chemoreceptors
-
Elongated transmembrane proteins in which binding to a ligand molecule induces a conformational change that affects downstream pathway activity.
- Trade-off
-
A situation in which a certain trait cannot increase without a decrease in another trait because of certain physical or biological constraints. When the constraint is lifted, the trade-off disappears.
- Adaptation time
-
The time required for the pathway activity and tumble bias to restore to pre-stimulus levels after prolonged stimulation.
- Phenotypic diversity
-
The variation in the biological traits among members of an isogenic population due to biochemical noise.
- Tumble bias
-
The relative proportion of time that a bacterium spends reorienting during motility. Cells with high tumble bias reorient more frequently.
- Pathway gain
-
How strongly a cell amplifies the signal from a given chemical gradient. The amplification is determined by the properties of the signal transduction machinery.
- Effective diffusivity
-
The rate at which a randomly swimming cell explores space.
- Brownian random walk
-
A type of random walk of small particles in a fluid, driven by thermal effects, which results in diffusive behaviour.
- Lévy flight
-
A type of superdiffusive random walk in which the step-length distribution is heavy tailed, leading to increased spatial exploration compared with Brownian motion.
- Allee effect
-
A positive density dependence of individual fitness which arises from cooperation or facilitation among individuals in the population.
- Glycolytic
-
Using the glycolysis pathway to generate energy.
Rights and permissions
About this article
Cite this article
Keegstra, J.M., Carrara, F. & Stocker, R. The ecological roles of bacterial chemotaxis. Nat Rev Microbiol 20, 491–504 (2022). https://doi.org/10.1038/s41579-022-00709-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-022-00709-w
This article is cited by
-
Spatial structure, chemotaxis and quorum sensing shape bacterial biomass accumulation in complex porous media
Nature Communications (2024)
-
Host association and intracellularity evolved multiple times independently in the Rickettsiales
Nature Communications (2024)
-
A Novel Device and Method for Assay of Bacterial Chemotaxis Towards Chemoattractants
Indian Journal of Microbiology (2024)
-
Green Waste Compost Impacts Microbial Functions Related to Carbohydrate Use and Active Dispersal in Plant Pathogen-Infested Soil
Microbial Ecology (2024)
-
Successional action of Bacteroidota and Firmicutes in decomposing straw polymers in a paddy soil
Environmental Microbiome (2023)