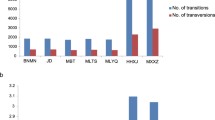
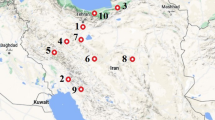
Abstract
Platostoma palustre (syn Mesona chinensis) (Lamiaceae) is an herbal plant used in traditional medicine and as food. During domestication over centuries, the plants for cultivation were propagated vegetatively by stem cuttings. However, the consequence of long-term clonal reproduction on genetic diversity and genetic structure is unknown for this plant. In this study, we used specific-locus amplified fragment sequencing (SLAF-seq) to identify genetic variation among 36 P. palustre populations collected from southern China. In total, 16,651 reliable single-nucleotide polymorphisms were obtained. A low level of genetic diversity, in the form of heterozygosity, was found at both the species level (expected heterozygosity, He = 0.207) and the population level (He = 0.023–0.162), indicating a narrow genetic base for the crop. This crop plant was initially selected over centuries from wild populations in southern China; Its domestication history, as well as its vegetative mode of propagation, may explain the low level of genetic diversity. On the other hand, values for pairwise fixation index (0.483) and analysis of the molecular variance (79.18%) indicated high genetic differentiation among populations. STRUCTURE analysis and phylogenetic tree analysis showed that the 36 populations studied could be divided into four groups. The initial differences in the germplasm introduced into the cultivated P. palustre populations from local wild resources and the limited gene flow among populations because of clonal propagation may have played important roles in shaping the pattern of genetic variation. The results will be helpful in the conservation of germplasm resources and in future breeding work on P. palustre.



Similar content being viewed by others
Availability of data and material
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1(5):2320–2325. https://doi.org/10.1038/nprot.2006.384
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32(3):314–331
Chang Y, Peng M, Chu G, Wang M (2020) Genome-wide assessment of population structure and genetic diversity for Anabasis aphylla based on specific length amplification fragment sequencing. J Plant Interact 15:75–82. https://doi.org/10.1080/17429145.2020.1750096
Chen JY, Liu P, Huang YZ, Su HL (2011) Analysis on genetic relationship of different provenances of Mesona chinensis Benth. with the ISSR technique. Chinese Wild Plant Res 30:46–50. https://doi.org/10.3969/j.issn.1006-9690.2011.05.011 (In Chinese with English abstract)
Chung J, Park Y (2010) Population structure analysis reveals the maintenance of isolated sub-populations of weedy rice. Weed Res 50:606–620. https://doi.org/10.1111/j.1365-3180.2010.00810.x
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
DeSilva R, Dodd RS (2020) Fragmented and isolated: limited gene flow coupled with weak isolation by environment in the paleoendemic giant sequoia (Sequoiadendron giganteum). Am J Bot 107:45–55. https://doi.org/10.1002/ajb2.1406
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Emerson KJ, Merz CR, Catchen JM, Hohenlohe PA, Cresko WA, Bradshaw WE, Holzapfel CM (2010) Resolving postglacial phylogeography using high-throughput sequencing. PNAS 107:16196–16200. https://doi.org/10.1073/pnas.1006538107
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fang H, Liu H, Ma R, Liu Y, Li J, Yu X, Zhang H, Yang Y, Zhang G (2020) Genome-wide assessment of population structure and genetic diversity of Chinese Lou onion using specific length amplified fragment (SLAF) sequencing. PLoS ONE 15(5):e231753. https://doi.org/10.1371/journal.pone.0231753
Gao Y, Yin S, Wu L, Dai D, Wang H, Liu C, Tang L (2017) Genetic diversity and structure of wild and cultivated Amorphophallus paeoniifolius populations in southwestern China as revealed by RAD-seq. Sci Rep-UK 7:14183. https://doi.org/10.1038/s41598-017-14738-6
Gevaert SD, Mandel JR, Burke JM, Donovan LA (2013) High genetic diversity and low population structure in Porter’s sunflower (Helianthus porteri). J Hered 3:407–415. https://doi.org/10.1093/jhered/est009
Goncalves-Oliveira RC, Wohrmann T, Benko-Iseppon AM, Krapp F, Alves M, Wanderley MDGL, Weising K (2017) Population genetic structure of the rock outcrop species Encholirium spectabile (Bromeliaceae): The role of pollination vs. seed dispersal and evolutionary implications. Am J Bot 104:868–878. https://doi.org/10.3732/ajb.1600410
Huang HC, Chuang SH, Wu YC, Chao PM (2016a) Hypolipidaemic function of Hsian-tsao tea (Mesona procumbens Hemsl.): Working mechanisms and active components. J Funct Foods 26:217–227. https://doi.org/10.1016/j.jff.2016.07.019
Huang L, Shen M, Zhang X, Jiang L, Song Q, Xie J (2018) Effect of high-pressure microfluidization treatment on the physicochemical properties and antioxidant activities of polysaccharide from Mesona chinensis Benth. Carbohyd Polym 200:191–199. https://doi.org/10.1016/j.carbpol.2018.07.087
Huang HR, Wu W, Zhang JX, Wang LJ, Yuan YM, Ge XJ (2016b) A genetic delineation of Patchouli (Pogostemon cablin) revealed by specific-locus amplified fragment sequencing. J Syst Evol 54:491–501. https://doi.org/10.1111/jse.12195
Keneni G, Bekele E, Imtiaz M, Dagne K, Getu E, Assefa F (2012) Genetic diversity and population structure of Ethiopian chickpea (Cicer arietinum L.) germplasm accessions from different geographical origins as revealed by microsatellite markers. Plant Mol Biol Rep 30:654–665. https://doi.org/10.1007/s11105-011-0374-6
Kent WJ (2002) BLAT – the BLAST-like alignment tool. Genome Res 12:656–664. http://www.genome.org/cgi/doi/https://doi.org/10.1101/gr.229202
Kishore G, Gupta S, Pandey A (2012) Assessment of population genetic diversity of Fagopyrum tataricum using SSR molecular markers. Biochem Syst Ecol 43:32–41. https://doi.org/10.1016/j.bse.2012.02.018
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a Dual-Index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microb 79:5112–5120. https://doi.org/10.1128/AEM.01043-13
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lai L, Chou S, Chao W (2001) Studies on the antioxidative activities of Hsian-tsao (Mesona procumbens Hemsl) leaf gum. J Agr Food Chem 49:963–968. https://doi.org/10.1021/jf001146k
Lai LS, Chao SJ (2000) Effects of salts on the thermal reversibility of starch and Hsian-tsao (Mesona procumbens Hemsl) leaf gum mixed system. J Food Sci 65:954–959. https://doi.org/10.1111/j.1365-2621.2000.tb09399.x
Li H, Geng S (2015) Assessment of population genetic diversity of Derris elliptica (Fabaceae) in China using microsatellite markers. Ind Crop Prod 73:9–15. https://doi.org/10.1016/j.indcrop.2015.04.023
Li Q, Li Y, Li Q, Chen Z, Chen J, Geng S (2021) Evaluation of morphological and phytochemical characteristics of Mesona chinensis populations in southern China. Plant Prod Sci 24(3):374–387. https://doi.org/10.1080/1343943X.2020.1847667
Liu XG, Chen MM (2004) Research on the explotation and utilization of Mesona Blume in China. Food Res Dev 25:109–112. https://doi.org/10.3969/j.issn.1005-6521.2004.05.040(In Chinese with English abstract)
Liu XG, Fang YP (1998) The development and utilization of Mesona chinensis Benth. Chinese Wild Plant Resour 17:29–32 (In Chinese)
Liu Z, Li H, Wen Z, Fan X, Li Y, Guan R, Guo Y, Wang S, Wang D, Qiu L (2017) Comparison of genetic diversity between Chinese and American soybean (Glycine max (L.)) accessions revealed by high-density SNPs. Front Plant Sci 8:2014. https://doi.org/10.3389/fpls.2017.02014
Liu S, Xiao Y, Shen M, Zhang X, Wang W, Xie J (2019) Effect of sodium carbonate on the gelation, rheology, texture and structural properties of maize starch-Mesona chinensis polysaccharide gel. Food Hydrocolloid 87:943–951. https://doi.org/10.1016/j.foodhyd.2018.09.025
Lyu Y, Dong X, Huang L, Zheng J, He X, Sun H, Jiang Z (2020) SLAF-seq uncovers the genetic diversity and adaptation of Chinese Elm (Ulmus parvifolia) in Eastern China. Forests 11:80. https://doi.org/10.3390/f11010080
McKey D, Elias M, Pujol B, Duputie A (2010) The evolutionary ecology of clonally propagated domesticated plants. New Phytol 186:318–332. https://doi.org/10.1111/j.1469-8137.2010.03210.x
Mengesha WA, Demissew S, Fay MF, Smith RJ, Nordal I, Wilkin P (2013) Genetic diversity and population structure of Guinea yams and their wild relatives in South and South West Ethiopia as revealed by microsatellite markers. Genet Resour Crop Evol 60:529–541. https://doi.org/10.1007/s10722-012-9856-0
Peakall R, Smouse P (2012) GenAlEx 6.5: Genetic analysis in Excel: Population genetic software for teaching and research- an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Sharma HK, Sarkar M, Choudhary SB, Kumar AA, Maruthi RT, Mitra J, Karmakar PG (2016) Diversity analysis based on agro-morphological traits and microsatellite based markers in global germplasm collections of roselle (Hibiscus sabdariffa L.). Ind Crop Prod 89:303–315. https://doi.org/10.1016/j.indcrop.2016.05.027
Shen J, Shan J, Zhu X, Xu Y, Li M (2020) Effect of Chinese herbal tea on health tested on Drosophila Model. Plant Food Hum Nutr 75:305–306. https://doi.org/10.1007/s11130-020-00809-z
Shen G, Sun M, Huang X, Wu Q (2000) The research and utilization of Mesona chinensis. Development of agriculture and animal husbandry products 2000:6–8. CNKI:SUN:LMKF.0.2000-05-001 (In Chinese)
Shrestha MK, Golan-Goldhirsh A, Ward D (2002) Population genetic structure and the conservation of isolated populations of Acacia raddiana in the Negev Desert. Biol Conserv 108:119–127. https://doi.org/10.1016/S0006-3207(02)00100-3
Shyu M, Kao T, Yen G (2008) Hsian-tsao (Mesona procumbens Heml.) prevents against rat liver fibrosis induced by CCl4 via inhibition of hepatic stellate cells activation. Food Chem Toxicol 46:3707–3713. https://doi.org/10.1016/j.fct.2008.09.051
Sun X, Liu D, Zhang X, Li W, Liu H, Hong W, Jiang C, Guan N, Ma C, Zeng H, Xu C, Song J, Huang L, Wang C, Shi J, Wang R, Zheng X, Lu C, Wang X, Zheng H (2013) SLAF-seq: An efficient method of large-scale De Novo SNP discovery and genotyping using High-Throughput Sequencing. PLoS ONE 8:e58700. https://doi.org/10.1371/journal.pone.0058700
Vicente M, Lopez C, Fulton T (2004) Genetic Diversity Analysis with Molecular Marker Data: Learning Module. International Plant Genetic Resources Institute (IPGRI), Rome, Italy
Warschefsky EJ, Wettberg EJBV (2019) Population genomic analysis of mango (Mangifera indica) suggests a complex history of domestication. New Phytol 222:2023–2037. https://doi.org/10.1111/nph.15731
Wei RC, Tan XM, Wu QH, Chen QP, Pan LM, Tang MQ, Li LX (2014) Standard operating procedure (SOP) for Mesona chinensis Benth. Jiangsu Agric Sci 42:198–200. https://doi.org/10.15889/j.issn.1002-1302.2014.02.217 (In Chinese)
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354. https://doi.org/10.1111/j.1469-1809.1949.tb02451.x
Wu QH (2011) The general situation of cultivation studies of Mesona chinensis Benth. North Hortic 2011:61–63. CNKI:SUN:BFYY.0.2011-24-022 (In Chinese with English abstract)
Wu TL (1995) Flora of Guangdong. In: Luo XR (ed) Lamiaceae. Guangdong Science and Technology Press, Guangzhou, pp 470–471
Wu Z, Raven H (1995) Flora of China. In: Li H, Hedge IC (1994) Lamiaceae. Science Press, Beijing, pp 297
Wurschum T, Langer SM, Longin CFH, Korzun V, Akhunov E, Ebmeyer E, Schachschneider R, Schacht J, Kazman E, Reif JC (2013) Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theor Appl Genet 126(6):1477–1486. https://doi.org/10.1007/s00122-013-2065-1
Xia W, Luo T, Zhang W, Mason AS, Huang D, Huang X, Tang W, Dou Y, Zhang C, Xiao Y (2019) Development of high-density SNP markers and their application in evaluating genetic diversity and population structure in Elaeis guineensis. Front Plant Sci 10:130. https://doi.org/10.3389/fpls.2019.00130
Zavinon F, Adoukonou-Sagbadja H, Keilwagen J, Lehnert H, Ordon F, Perovic D (2020) Genetic diversity and population structure in Beninese pigeon pea [Cajanus cajan (L.) Huth] landraces collection revealed by SSR and genome wide SNP markers. Genet Resour Crop Evol 67:191–208. https://doi.org/10.1007/s10722-019-00864-9
Zhang GF, Guan JM, Lai XP, Lin J, Liu JM, Xu HH (2012) RAPD fingerprint construction and genetic similarity of Mesona chinensis (Lamiaceae) in China. Genet Mol Res 11:3649–3657. https://doi.org/10.4238/2012.October.4.12
Zhong SH, Huang DQ (2007) The spatial distribution and group characteristic of Chinese Hakkas. Guangxi Ethnics Stud 4:80–85. https://doi.org/10.3969/j.issn.1004-454X.2007.04.012 (In Chinese with English abstract)
Acknowledgements
We would like to thank Yanping Li for helping with the collection of samples.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31670324); the Foundation of State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKL-CUSAb-2013–11); and the Foundation of Shenzhen Key Laboratory of Southern Subtropical Plant Diversity (SSTLAB-2013–03).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Qi Li wrote the manuscript and carried out bioinformatic analyses. Qi Li and Wei Xia extracted the DNA. Qi Li and Qian Li performed data collection. Xuezhi Liang, Jianguang Xie, and Shilei Geng collected the plant materials. Shilei Geng reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Q., Xia, W., Li, Q. et al. Genome-wide assessment of genetic diversity and population structure of Platostoma palustre in southern China and implications for conservation and breeding strategies. Genet Resour Crop Evol 69, 1349–1364 (2022). https://doi.org/10.1007/s10722-021-01308-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01308-z