Abstract
Chloroplast markers (cp markers) are the useful instrument for cannabis (syn. hemp, Cannabis sativa L.) to study relationships of accessions between different geographic origins. In an alignment of three published plastomes 38 chloroplast polymorphisms were identified from which 8 cp markers were used to study relationships of 53 cannabis accessions by high-resolution-melting analyis (HRMA). The marker set could distinguish six haplotypes (‘A’ to ‘F’) in the cannabis collection, where haplotypes ‘A’ and ‘F’ dominated with 34% and 50% of the individuals, respectively. A majority of populations (37) were homogeneous regarding the haplotype, 12 accessions were constituted of two haplotypes and 4 accessions of three haplotypes. Most of the European fibre cultivars consisted of the ‘F’-type (e.g. ‘Fibrimon’, Fibrimon 21’, Juso 14’, ‘Fasamo’ and ‘Schurig’), some were mixed ‘A/F’-types (e.g. ‘Fibrimon 21’, ‘Superfibra’, ‘Lorrin 110’, ‘Futura’, ‘Havelländische’). The Italian ‘Carmagnola in Selezione’ was exceptional in being a pure ‘A’-type. In the heterogenous populations, expected heterozygosity ranged from 0.06 to 0.41. The populations were well differentiated by this marker set locating 79% of the variation among populations (AMOVA). By comparison with plastomes from the closest related genus Humulus, haplotype ‘B’ could be identified as haplotype of the common ancestor of both genera. The haplotype ‘B’ is rare with a frequency of only 4% in the populations analysed. Unfortunately, the true geographic origin of most samples was unclear. However, amongst all published plastomes, only two were classified as haplotype ‘B’, both pointing independently back to Yunnan province (China), indicating Yunnan as the region of origin of the genus Cannabis.
Similar content being viewed by others
Introduction
Cannabis sativa L. (hemp or cannabis) belongs to the small family of Cannabaceae, which comprises of ten genera with Humulus (hop, 3 species) as sister genus (Yang et al. 2013). The genus Cannabis is monotypic (Small and Cronquist 1976) and it has been distributed globally as one of the oldest known crop plants. The use of C. sativa can be characterized as multi-purpose of fibre for paper, textile or construction materials (Karus and Vogt 2004), as seeds for food and feed (Callaway 2004) and the female inflorescence as medicine (Ben Amar 2006), and psychotropic drug (Szendrei 1998).
Central Asia is regarded as origin of C. sativa (de Candolle 1885; McPartland et al. 2018). According to McKim (2003), the first archaeological discoveries in China are from the Neolithic period, around 4000 BC, while Long et al. (2017) date the first utilization to 8,000 BC. In contrast to the prevalent Central-Asia-Origin hypothesis of C. sativa, molecular evidence reveals that this species probably comes from a low latitude region of India (Zhang et al. 2018a). The most profound localisation of the origin was addressed by McPartland et al. (2018), who identified the northeastern Tibetan plateau near Qinquai Lake as origin by pollen analysis. From there, C. sativa spread over 6 Ma ago to Europe, 1.2 Ma ago to eastern China and 32.6 thousand years ago to India. C. sativa favors a mild climate with sufficient water and sunlight, and early humans spread it into a range of favorable temperate and sub-tropical niches where it becomes naturalized throughout Eurasia, in parts of Africa, and more recently in the New World (Clarke and Merlin 2016).
Molecular plant phylogeographic studies have mostly relied on the chloroplast (cp) genome because of the low mutation rate of this single and non-recombining unit of inheritance (Schaal et al. 1998). In C. sativa, cpDNA has been ascertained as a valuable tool for such analysis with sufficient variability on the interpopulational level (Gilmore et al. 2007; Zhang et al. 2018a).
Genebank collections are composed of genetic materials with original sources from in situ conditions or from breeding/research programs. Many of these materials can no longer be found in situ for a variety of reasons (Fowler and Hodgkin 2004). Gene banks were created at the beginning of the twentieth century as repositories of genetic material to preserve genetic diversity and to provide easy access of genetic materials to breeders (Fowler and Hodgkin 2004). The difficulty of maintaining such ressources for medicinal plants is their uncountable number of taxa (Lohwasser and Weise 2020).
This study develops molecular markers from cpDNA of C. sativa for analyzing the variability and relationsships of cannabis accessions of the genebank Gatersleben and published plastomes.
Material and methods
Development of chloroplast SNP markers
The chloroplast genomes of three C. sativa genotypes (Genbank ID/cultivar: KR363961/‘Yoruba Nigeria’ (Oh et al. 2016), KP274871/‘Carmagnola’, KR779995/‘Dagestani’ (Matielo et al. 2020)) were assembled, SNPs localised and primers for SNP candidates developed with Primer3 (Untergasser et al. 2012) as implemented in Geneious Prime 2020.2.2 (Biomatters Ltd.). The SNP candidates were tested with a test set of 33 individuals of different accessions with high-resolution melting analysis. Of the 29 SNP candidates, 8 markers were selected on the basis of curve types easy to distinguish. Two individuals representing the two curve types per marker were selected and furtheron added as references to each run.
Sample material
Fifty-three accessions of the IPK collection were grown in the greenhouse and leaves from 10 plants per accession were sampled with the exception of CAN33 and CAN60, where only 9 and 8 plants could be sampled, respectively. Therefore, the study was comprised of 527 Cannabis individuals. The leaf samples were dried at 38 °C in a drying oven and stored until analysis.
DNA Extraction, PCR and HRM
DNA was extracted with a modified CTAB method (Schmiderer et al. 2013). Concentration and quality of the DNA were determined on a 1.5% agarose gel electrophoresis and a NanoDrop 2000 (Fisher Scientific). HRM with pre-amplification was performed on a Rotor-Gene 6000 (Qiagen). For a PCR reaction in 10 µl, 1 µl of genomic DNA (1:100 dilutions of the original DNA extract) was added to a master mix containing 1 × HOT FIREPol® EvaGreen® HRM Mix (no ROX) (Solis BioDyne) and 100 nM forward and reverse primers (ordered at Life Technologies), respectively. The PCR cycle profile included a denaturation step at 95 °C for 14 min, followed by 45 cycles (95 °C for 10 s, 59 °C for 20 s and 72 °C for 20 s) with a final denaturation step at 95 °C for 30 s. For high-resolution melting curve analysis (HRM) the temperature was increased from 69 °C to 81 °C by 0.1 °C/s. All reactions were completed in duplicates with non-target controls in each run.
Statistical analysis
The statistical analyses were done with R 3.6.2. (R Core Team 2019) under Rstudio 1.2.5033 (RStudio Team 2019) using the packages poppr (Kamvar et al. 2014, 2015) and ggtree (Yu et al. 2017). Distances were calculated according to Prevosti et al. (1975), the Simpson index (Simpson 1949), Nei’s expected heterozygosity (HEXP) (Nei 1978), as well as the genetic differentiation using GST (Hedrick 2005). As a measure of linkage disequilibrium, \({\bar{r}}\)d (an adapted form of the index of association IA (Brown et al. 1980)) were calculated (Agapow and Burt 2001).
The identified haplotypes were compared with Geneious Prime 2020.2.2 (Biomatters Ltd.) to complete chloroplasts of hop species (Humulus lupulus (KT266264, MG573060), H. scandens (MH118122) and H. yunnanensis (MK423880)) and cannabis (C. sativa (KY084475), C. sativa cv. ‘Carmagnola’ (NC026562), C. sativa cv. ‘Dagestani’ (KR779995), C. sativa cv. ‘Yoruba Nigeria’ (NC027223), C. sativa cv. ‘Yunma 7’ (MW013540), C. sativa (MH118118) and C. sativa cv. ‘Cheungsam’ (KR184827)).
Results
Marker development
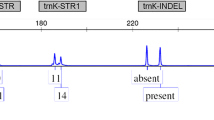
Fifty-three accessions of C. sativa from the genebank Gatersleben were analysed with 8 chloroplast markers using high-resolution melting analysis (HRM). In order to detect chloroplast markers, three published chloroplast genomes of C. sativa were aligned and 38 polymorphisms (16 indels and 16 SNP, of which 7 transitions) were identified (Supplementary Table 1). Candidates were preselected on their theoretical suitability for high-resolution melting analysis (cf. exemplarily to Supplementary Fig. 1). Those candidates were evaluated with a small sample set and then narrowed to a set of one INDEL (marker P18) and seven SNP’s (Tables 1, 2). As in many plant species, cannabis chloroplast DNA contains two inverted repeats (26,011 bp each), which separate a large single copy region (84,059 bp) from a small single copy region (17,829 bp) (Zhang et al. 2018b). All but one markers were located in the large single copy region, only marker P10 was in the small single copy region. Five markers were intergenic, one (P21) in an intron of rps16, and two in a coding region (P18 in matK-trnK-UUU and P12 in rps11) (Table 1).
Description of the markers
The expected heterozygosity of the markers over all populations was in average 0.37 with most markers in a narrow range between 0.45 and 0.5 (Table 2) while two markers were very low with 0.063 (P26) and 0.077 (P10). The average GST of all markers was 0.87 with a GST of P26 with 0.76 and P10 with 0.96 as two extremes, while all other markers ranged between 0.87 and 0.91.
Haplotypes
In total, 6 haplotypes could be identified, denominated haplotypes ‘A’ to ‘F’ (Figs. 1, 2). 84% of the individuals were classified either as haplotype ‘A’ (34%) or ‘F’ (50%). All other haplotypes are filling successively the gap between the two most extreme haplotypes ‘A’ and ‘F’ (‘B’: 4%, ‘C’: 1%, ‘D’: 3% and ‘E’: 8%). Thirty-seven accessions (70%) were homogeneous with only one haplotype (Fig. 1). Out of the 37 homogeneous populations, 21 were pure ‘F’, 10 pure ‘A’, 3 pure ‘E’, 2 pure ‘B’ and 1 pure ‘D’. The haplotype ‘C’ was only present in mixed accessions. In the heterogenous accessions, 12 were found with two haplotypes and four with three haplotypes. Most of the heterogenous populations (9 out of 12 the populations with two haplotypes) contained the haplotypes ‘A’ and ‘F’.
No geographical pattern of haplotypes could be observed from the accessions’ passport data (data not shown). However, the most commonly known European fibre type cultivars consisted of haplotype ‘F’ or ‘F’ mixed with another haplotype (in most cases mixed with the ‘A’-type). For ‘Fibrimon’ and ‘Kompolti’, three accessions per cultivar from different providers were in our sample set. All ‘Fibrimon’ accessions were pure ‘F’ haplotype, while two accessions of cv. ‘Kompolti’ were ‘F’-type and one a mixed ‘A/F’-type. Other fibre cultivars in the genebank could also be grouped in either pure ‘F’-type or mixed ‘A/F’-type. Pure ‘F’-types were ‘Fibrimon 21’, Juso 14’, ‘Fasamo’ and ‘Schurig’. Mixed ‘A/F’-type were ‘Fibrimon 56’, ‘Eletta Campana’, ‘Superfibra’, ‘Lorrin 110’, ‘Futura’ and ‘Havelländische’. The Italian ‘Carmagnola in Selezione’ was – as a singular exception amongst the fibre accessions—a pure ‘A’-type.
Basic populations statistics were calculated for the heterogenous populations separately and for all populations as an overall mean (Table 3). The Shannon–Wiener index of haplotype diversity ranged in the heterogenous populations from 0.33 to 0.944 and the Simpson Index from 0.18 to 0.56. A number of mixed populations had only one individual of a different haplotype (evenness = 0.57). In only one population (52) the number of individuals of different haplotypes was in balance (evenness = 1). The expected heterozygosity ranged from 0.06 to 0.41. The level of linkage disequilibrium was in all heterogenous populations highly significant and ranged from 0.58 to 1. Overall (so including all individuals), the Shannon–Wiener index was 1.19, Simpson’s index 0.63, the evenness 0.73, the expected heterozygosity 0.37 and the index of association 0.53. In the analysis of molecular variance (AMOVA), the populations were well differentiated with 79% of the variation located among populations Table 4).
All three Humulus species with published plastomes (H. lupulus, H. scandens and H. yunnanensis), as well as C. sativa (Herbarium of the Kunming Institute of Botany, province Yunnan, China (Zhang et al. 2018b) and the cannabis fibre variety ‘Yunma 7’ (Deng et al. 2021)) were characterized as haplotype ‘B’. Therefore, haplotype ‘B’ can undoubtedly be regarded as the ancient haplotype where the other haplotypes in cannabis were evolved from (Fig. 2). None of the cannabis polymorphisms were polymorphic in the hop chloroplasts, all mutations were occurring in Cannabis after separation of the two genera from their common ancestor. Two genebank accessions in our study were belonging to haplotype ‘B’, one from France (no further background information available) and one from Spain designated as ‘Kongo Hanf’ (= ‘Congo hemp’). Two of the published cannabis plastomes were also belonging to haplotype 'B', (cv. 'Yunma 7' and an accession without any further description, submitted by the Kunming Institute of Botany, Yunnan, China (GenBank accession no. MH118118)). Further published cannabis plastomes were haplotypes ‘A’ (cv. ‘Carmagnola’), ‘C’ (cv. ‘Cheongsam’) and ‘F’ (cv. ‘Yoruba Nigeria’ and cv. ‘Dagestani’).
Discussion
Marker development
Chloroplast markers have several advantages, such as maternal inheritance. As a result, they are usually useful to explore genetic structure and gene flow between rather than within populations, field of applications are evolutionary studies, migration of plants (biogeography) and profiling genotypes and gene pools. The highly significant linkage disequilibrium, that are usually in plants used to identify clonality, could have been also expected for non-recombining maternal lineages as in chloroplasts or mitochondria.
Also in C. sativa, nuclear marker variability (e.g. expected heterozygosities of 0.68 (Gilmore and Peakall 2002) or 0.75 (Soler et al. 2017)) was higher than chloroplast variability (expected heterozygosity of 0.37 in our study). However, one study showed expected heterozygosities of nuclear markers below that of our results (0.22 to 0.32 (Lynch et al. 2016)).
Nuclear markers revealed higher intrapopulational cannabis variability, demonstrated with microsatellite markers which attributed only 32% to the variation between cultivars, while 37% and 31% was intra-cultivar and intra-individual, respectively (Soler et al. 2017). Chloroplast markers moved the focus to the higher level of between cultivar variability with 69% (Zhang et al. 2018a) to 79% (this study). The major disadvantage of cp (or mt) markers are their limitations in absolute numbers, 38 cp markers in total based on three cannabis chloroplast genomes in comparison to (fractional) 24,710 ncSNPs (Soorni et al. 2017) or 14,031 ncSNPs (Sawler et al. 2015). However, depending on the type of query, just a few, but powerful cp markers may deliver sufficient information for an intra-specific classification e.g. for identifying different genepools (Gilmore and Peakall 2002).
Overlapping accessions in different cpDNA studies
Gilmore et al. (2007), developed 5 cpDNA and 2 mtDNA markers with good discrimination power of accessions and identified with this set 6 haplotypes that clustered the samples into three haplotype groups. Comparing some jointly used fibre cultivars showed that the grouping of Gilmore et al. (2007) was not the same as ours. Zhang et al. (2018a) sequenced with 5 primer pairs in highly variable cp regions and identified 23 haplotypes that grouped nicely into 3 haplotype groups. Overlapping accessions exhibited that our haplotypes ‘C’ and ‘F’ could be found in haplogroup ‘H’ in the study of Zhang et al. (2018a) and our haplotype ‘A’ fell into haplogroup ‘M’. This could be evident that haplotype ‘A’ was already in Central and Western China, while haplotype ‘F’ is found in North-East and North-West of China. Studies of Mongolian cannabis accession could possibly bridge the two distribution areas of haplogroup ‘M’/haplotype ‘F’, providing strong evidence of the origin of this haplogroup/haplotype already in todays Northern China or North of China, followed by a Western migration to Europe. This mutational differentiation of the chloroplast in the North was probably accompanied by ecological adaptation to Northern conditions (e.g. day length, temperature) that made the ‘F’-type so successful in migrating west. This example of finding common ground between studies shows that harmonizing cannabis cpSNP would create an effective tool in the future.
Is the origin of cannabis in the Chinese province Yunnan?
The haplotype ‘B’ is common in Cannabis and Humulus and must have been present in the common ancestor of the two genera. The identification of the original haplotype allowed the determination of the sequence of the eight cp mutations used here over time because of the non-recombining maternal lineages. Cannabis and Humulus diverged between 18.23 mya (8.83–36.56 mya) (Zhang et al. 2018a) and 27.8 mya (McPartland 2018). Pollen analysis pinned the center of Cannabis origin to the northeastern Tibetan plateau near Qinquai Lake from where it spreaded west to Europe (6 mya ago), east to eastern China (1.2 mya ago) and to India (only 32.6 thousand years ago) (McPartland et al. 2018). The spreading was supported by the estimation of the crown age of Cannabis to 2.24 mya (0.81–5.81 mya) (Zhang et al. 2018a). The two ‘B’-type accessions submitted to the genebank Gatersleben from France (no further information provided) and Spain (‘Congo hemp’) were not helpful in tracing back the origin of cannabis. More informative were the two published plastomes with haplotype ‘B’ from independent sources, a plastome from a plant of the Herbarium of the Kunming Institute of Botany (province Yunnan, China) (Zhang et al. 2018b) and the plastome of cv. ‘Yunma 7’, a main cultivar in fibre production (Deng et al. 2021), bred in Yunnan province (Amaducci et al. 2015). This province has a long tradition of using cannabis (Clarke and Gu 1998) and is one of the main production areas of hemp fibre in China (Deng et al. 2021). Provided that both samples were originally collected from natural stands in Yunnan, cannabis could have had its origin in this province.
Cp haplotypes in the European breeding history for fibre use
In Europe, domestication of Cannabis was occurring in the copper/bronze age indicative of an domestication event independent from the Chinese domestication (McPartland et al. 2017). Most European fibre cultivars were derived from European landraces and consisted—at least partly—of haplotype ‘F’, corresponding to the ‘fibre-type’ haplotype ‘1,122,121’ of Gilmore et al. (2007). The monoecious German cultivar ‘Fibrimon’ (three accessions in the genebank, all ‘F’-type accession) was found to be bred from old German origins, probably landraces, namely ‘Schurig’ (‘F’-type) and ‘Havelländer’ (‘A/F’-type), both originally of Central-Russian origin (Hoffmann 1961). Since monoecisms is a desired trait for fibre use, but rare in C. sativa, most of the French cultivars (e.g. ‘Fibrimon 21’ (‘F’-type) and ‘Fibrimon 56’ (‘A/F’-type) go back to ‘Fibrimon’ (de Meijer 1995). The Hungarian variety ‘Kompolti’ (three genebank accessions, two ‘F’-type, one ‘A/F’-type) was obtained from ‘Fleischmann hemp’ which had its origin in Italy (de Meijer 1995). The Romanian cultivar ‘Lovrin 110’ (‘A/F’-type) was derived from Bulgarian landraces and the Russian ‘Juso 14’ (‘F’-type) from ‘JUS-6’, a crossing between a Southern origin, a Northern Russian dwarf origin, and the German ‘Odnodomnaya Bernburga’ (de Meijer 1995). The Italian ‘Eletta Campana’ (‘A/F’-type) originated from a selection from a Northern Italian landrace ‘Carmagnola’ and high fibre strains from Germany (de Meijer 1995). In breeding, selection itself is not restricted to geographical distinct entities, but includes all materials useful and approachable. Chinese fibre strains e.g. were the basis for some fibre cultivars in the United States at the beginning of the twentieth century. The cultivar ‘Chington’ (China-Washington), extensively used by hempseed growers in Kentucky, was developed from seeds obtained from Hankow, China (Dewey 1927). The Hungarian three-way hybrid ‘Kompolti Hybrid TC’ had also a Chinese component (de Meijer 1995). So, many cultivars used today may be based on already early global exchange explaining the occurrence of the ‘A’-type in fibre cultivars.
Accessions in genebanks are a mixture of donations from many different sources, genebank exchanges and own collection trips. Collection trips can usually be regarded as only trustful sources when it comes to a defined geographic origin, since donations are in most cases of selected materials, collected from mostly undefined sources planted in a field and often pollinated without isolation. That is demonstrated by the variety ‘Kompolti’, present in the genebank from three different donors. ‘Kompolti’ is usually a pure ‘F’-type. One accession, however, was a mixture of the haplotypes ‘F’ and ‘A’. Since chloroplasts are only maternally inherited, accidential cross-pollination can be ruled out. Such a mixture can only origin by mixing seeds. For cannabis, even collection trips were often not reliable sources due to the exchange of genetic materials for hundreds of years and over long distances and subsequent subspontaneous naturalization, either unintentionally or by cultivation of illegitimate strains hidden in a natural environment (Szendrei 1998). Therefore, it is difficult to distinguish natural from naturalized populations in cannabis.
Data availability
Data and DNA available on reasonable request.
References
Agapow P-M, Burt A (2001) Indices of multilocus linkage disequilibrium. Mol Ecol Notes 1:101–102. https://doi.org/10.1046/j.1471-8278.2000.00014.x
Amaducci S, Scordia D, Liu FH, Zhang Q, Guo H, Testa G, Cosentino SL (2015) Key cultivation techniques for hemp in Europe and China. Ind Crops Prod 68:2–16. https://doi.org/10.1016/j.indcrop.2014.06.041
Ben Amar M (2006) Cannabinoids in medicine: a review of their therapeutic potential. J Ethnopharmacol 105:1–25. https://doi.org/10.1016/j.jep.2006.02.001
Brown AH, Feldman MW, Nevo E (1980) Multilocus structure of natural populations of Hordeum spontaneum . Genetics 96:523–536. https://doi.org/10.1093/genetics/96.2.523
Callaway JC (2004) Hempseed as a nutritional resource: an overview. Euphytica 140:65–72. https://doi.org/10.1007/s10681-004-4811-6
Clarke RC, Gu W (1998) Survey of hemp (Cannabis sativa L.) use by the Hmong (Miao) of the Cina/Vietnam border region. J Int Hemp Assoc 5:4–9
Clarke RC, Merlin MD (2016) Cannabis domestication, breeding history, present-day genetic diversity, and future prospects. Crit Rev Plant Sci 35:293–327. https://doi.org/10.1080/07352689.2016.1267498
de Candolle A (1885) Origin of cultivated plants. New York, D. Appleton
de Meijer E (1995) Fibre hemp cultivars: A survey of origin, ancestry, availability and brief agronomic characteristics. J Int Hemp Assoc 66–73
Deng G, Yang M, Zhao K, Yang Y, Huang X, Cheng X (2021) The complete chloroplast genome of Cannabis sativa variety Yunma 7. Mitochondrial DNA B Resour 6:531–532. https://doi.org/10.1080/23802359.2021.1873709
Dewey LH (1927) Hemp varieties of improved type are result of selection. Yearbook of the USDA 358–361
Fowler C, Hodgkin T (2004) Plant genetic resources for food and agriculture: assessing Global availability. Annu Rev Environ Resour 29:143–179. https://doi.org/10.1146/annurev.energy.29.062403.102203
Gilmore S, Peakall R (2002) Isolation of microsatellite markers in Cannabis sativa L. (marijuana). Mol Ecol Notes 2003:105–107. https://doi.org/10.1046/j.1471-8286.2003.00367.x
Gilmore S, Peakall R, Robertson J (2007) Organelle DNA haplotypes reflect crop-use characteristics and geographic origins of Cannabis sativa. Forensic Sci Int 172:179–190. https://doi.org/10.1016/j.forsciint.2006.10.025
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638. https://doi.org/10.1111/j.0014-3820.2005.tb01814.x
Hoffmann W (1961) Hanf, Cannabis sativa L. In: Kappert H, Rudorf W (eds) Handbuch der Pflanzenzüchtung, V. Paul Parey, Berlin, pp 204–261
Kamvar ZN, Tabima JF, Gruenwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. https://doi.org/10.7717/peerj.281
Kamvar ZN, Brooks JC, Gruenwald NJ (2015) Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet 6:208. https://doi.org/10.3389/fgene.2015.00208
Karus M, Vogt D (2004) European hemp industry: cultivation, processing and product lines. Euphytica 140:7–12. https://doi.org/10.1007/s10681-004-4810-7
Lohwasser U, Weise S (2020) Genetic resources of medicinal and aromatic plants. In: Novak J, Blüthner W-D (eds) Medicinal, aromatic and stimulant plants. Springer, Cham, pp 1–205
Long T, Wagner M, Demske D, Leipe C, Tarasov PE (2017) Cannabis in Eurasia: origin of human use and bronze age trans-continental connections. Veget Hist Archaeobot 26:245–258. https://doi.org/10.1007/s00334-016-0579-6
Lynch RC, Vergara D, Tittes S, White K, Schwartz CJ, Gibbs MJ, Ruthenburg TC, deCesare K, Land DP, Kane NC (2016) Genomic and chemical diversity in cannabis. Crit Rev Plant Sci 35:349–363. https://doi.org/10.1080/07352689.2016.1265363
Matielo CBD, Lemos RPM, Sarzi DS, Machado LdO, Beise DC, Dobbler PCT, Castro RM, Fett MS, Roesch LFW, Camargo FAdO, Stefenon VM (2020) Whole plastome sequences of two drug-type cannabis: insights into the use of plastid in forensic analyses. J Forensic Sci 65:259–265. https://doi.org/10.1111/1556-4029.14155
McKim WA (2003) Drugs and behavior: an introduction to behavioral pharmacology, 5th edn. Prentice Hall, Upper Saddle River, N.J.
McPartland JM (2018) Cannabis systematics at the levels of family, genus, and species. Cannabis Cannabinoid Res 3:203–212. https://doi.org/10.1089/can.2018.0039
McPartland JM, Guy GW, Hegman W (2017) Cannabis is indigenous to Europe and cultivation began during the Copper or Bronze age: a probabilistic synthesis of fossil pollen studies. Veg Hist Archaeobot 2018:635–648. https://doi.org/10.1007/s00334-018-0678-7
McPartland JM, Hegman W, Long T (2018) Cannabis in Asia: its center of origin and early cultivation, based on a synthesis of subfossil pollen and archaeobotanical studies. Veget History Archaeobot. https://doi.org/10.1007/s00334-019-00731-8
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Oh H, Seo B, Lee S, Ahn D-H, Jo E, Park J-K, Min G-S (2016) Two complete chloroplast genome sequences of Cannabis sativa varieties. Mitochondrial DNA A DNA Mapp Seq Anal 27:2835–2837. https://doi.org/10.3109/19401736.2015.1053117
Prevosti A, Ocaña J, Alonso G (1975) Distances between populations of Drosophila subobscura, based on chromosome arrangement frequencies. Theor Appl Genet 45:231–241. https://doi.org/10.1007/BF00831894
R Core Team (2019) R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria
RStudio Team (2019) RStudio. RStudio, Inc., Boston, MA. www.rstudio.com
SantaLucia J (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A 95:1460–1465. https://doi.org/10.1073/pnas.95.4.1460
Sawler J, Stout JM, Gardner KM, Hudson D, Vidmar J, Butler L, Page JE, Myles S (2015) The genetic structure of marijuana and hemp. PLoS ONE 10:e0133292. https://doi.org/10.1371/journal.pone.0133292
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Mol Ecol 7:465–474. https://doi.org/10.1046/j.1365-294x.1998.00318.x
Schmiderer C, Lukas B, Novak J (2013) Effect of different DNA extraction methods and DNA dilutions on the amplification success in the PCR of different medicinal and aromatic plants. J Med Spice Plants 18:65–72
Simpson E (1949) Measurement of diversity. Nature 163:688. https://doi.org/10.1038/163688a0
Small E, Cronquist A (1976) A practical and natural taxonomy for Cannabis. Taxon 25:405–435. https://doi.org/10.2307/1220524
Soler S, Gramazio P, Figàs MR, Vilanova S, Rosa E, Llosa ER, Borràs D, Plazas M, Prohens J (2017) Genetic structure of Cannabis sativa var. indica cultivars based on genomic SSR (gSSR) markers: implications for breeding and germplasm management. Ind Crops Prod 104:171–178. https://doi.org/10.1016/j.indcrop.2017.04.043
Soorni A, Fatahi R, Haak DC, Salami SA, Bombarely A (2017) Assessment of genetic diversity and population structure in iranian cannabis germplasm.https://doi.org/10.1038/s41598-017-15816-5
Szendrei K (1998) Cannabis as an illicit narcotic crop: a review of the global situation of cannabis consumption, trafficking and production. Bull Narc 50:45–83
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. https://doi.org/10.1093/nar/gks596
Yang M-Q, van Velzen R, Bakker FT, Sattarian A, Li D-Z, Yi T-S (2013) Molecular phylogenetics and character evolution of Cannabaceae. Taxon 62:473–485. https://doi.org/10.12705/623.9
Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y (2017) ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. https://doi.org/10.1111/2041-210X.12628
Zhang Q, Chen X, Guo H, Trindade L, Salentijn E, Guo R, Guo M, Xu Y, Yang M (2018a) Latitudinal adaption and genetic insights into the origins of Cannabis sativa L. Front Plant Sci. https://doi.org/10.3389/fpls.2018.01876
Zhang H, Jin J, Moore MJ, Yi T, Li D (2018b) Plastome characteristics of Cannabaceae. Plant Divers 40:127–137. https://doi.org/10.1016/j.pld.2018.04.003
Acknowledgements
We cordially thank Jonathan Mayer for his great support.
Funding
Open access funding provided by University of Veterinary Medicine Vienna. No funding received.
Author information
Authors and Affiliations
Contributions
EO laboratory analysis, data evaluation, manuscript preparation; UL concept planning, statistical evaluation, seed bank information, manuscript; DJ laboratory analysis, development of markers, manuscript (technical issues); JR planning, marker development, assay optimisation, data evaluation, manuscript; JN concept, planning, statistical evaluation, manuscript preparation.
Corresponding author
Ethics declarations
Conflicts of interest
No conflicts of interests or competing interests.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Osterberger, E., Lohwasser, U., Jovanovic, D. et al. The origin of the genus Cannabis. Genet Resour Crop Evol 69, 1439–1449 (2022). https://doi.org/10.1007/s10722-021-01309-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01309-y