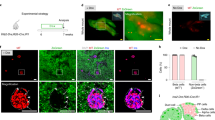
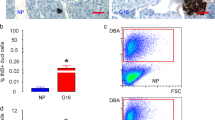
Abstract
Generation of beta cells via transdifferentiation of other cell types is a promising avenue for the treatment of diabetes. Here we reconstruct a single-cell atlas of the human fetal and neonatal small intestine. We identify a subset of fetal enteroendocrine K/L cells that express high levels of insulin and other beta cell genes. Our findings highlight a potential extra-pancreatic source of beta cells and expose its molecular blueprint.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data generated in this study are available at the Zenodo repository: https://doi.org/10.5281/zenodo.5457926. Immune cells from fetal samples 1100A and 1102 and the neonatal sample 1127 were presented in Olaloye et al.36. Data from Cao et al.8 are available at https://descartes.brotmanbaty.org/bbi/human-gene-expression-during-development/. Data from Baron et al.9 are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84133. Data from Elmentaite et al.14 are available at https://www.gutcellatlas.org/.
Code availability
All custom code used in this study is available at the Zenodo repository: https://doi.org/10.5281/zenodo.5457926.
References
Merrell, A. J. & Stanger, B. Z. Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat. Rev. Mol. Cell Biol. 17, 413–425 (2016).
Jennings, R. E., Berry, A. A., Strutt, J. P., Gerrard, D. T. & Hanley, N. A. Human pancreas development. Development 142, 3126–3137 (2015).
Habib, A. M. et al. Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry. Endocrinology 153, 3054–3065 (2012).
Talchai, C., Xuan, S., Kitamura, T., DePinho, R. A. & Accili, D. Generation of functional insulin-producing cells in the gut by Foxo1 ablation. Nat. Genet. 44, 406–412 (2012).
Bouchi, R. et al. FOXO1 inhibition yields functional insulin-producing cells in human gut organoid cultures. Nat. Commun. 5, 4242 (2014).
Chen, Y.-J. et al. De novo formation of insulin-producing ‘neo-β cell islets’ from intestinal crypts. Cell Rep. 6, 1046–1058 (2014).
Suissa, Y. et al. Gastrin: a distinct fate of neurogenin3 positive progenitor cells in the embryonic pancreas. PLoS ONE 8, e70397 (2013).
Cao, J. et al. A human cell atlas of fetal gene expression. Science 370, eaba7721 (2020).
Baron, M. et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360 (2016).
Blum, B. et al. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat. Biotechnol. 30, 261–264 (2012).
Matschinsky, F. M. & Wilson, D. F. The central role of glucokinase in glucose homeostasis: a perspective 50 years after demonstrating the presence of the enzyme in islets of Langerhans. Front. Physiol. 10, 148 (2019).
Vivot, K. et al. The regulator of G-protein signaling RGS16 promotes insulin secretion and β-cell proliferation in rodent and human islets. Mol. Metab. 5, 988–996 (2016).
Gross, S. et al. Lmx1a functions in intestinal serotonin-producing enterochromaffin cells downstream of Nkx2.2. Development 143, 2616–2628 (2016).
Elmentaite, R. et al. Single-cell sequencing of developing human gut reveals transcriptional links to childhood Crohn’s disease. Dev. Cell 55, 771–783 (2020).
Farack, L. et al. Transcriptional heterogeneity of beta cells in the intact pancreas. Dev. Cell 48, 115–125 (2019).
Nowak-Sliwinska, P. et al. Oncofoetal insulin receptor isoform A marks the tumour endothelium; an underestimated pathway during tumour angiogenesis and angiostatic treatment. Br. J. Cancer 120, 218–228 (2019).
Menon, R. K. & Sperling, M. A. Insulin as a growth factor. Endocrinol. Metab. Clin. North Am. 25, 633–647 (1996).
Herrera, P. L. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 127, 2317–2322 (2000).
Teitelman, G., Alpert, S., Polak, J. M., Martinez, A. & Hanahan, D. Precursor cells of mouse endocrine pancreas coexpress insulin, glucagon and the neuronal proteins tyrosine hydroxylase and neuropeptide Y, but not pancreatic polypeptide. Development 118, 1031–1039 (1993).
Kaspi, H., Pasvolsky, R. & Hornstein, E. Could microRNAs contribute to the maintenance of β cell identity? Trends Endocrinol. Metab. 25, 285–292 (2014).
Stras, S. F. et al. Maturation of the human intestinal immune system occurs early in fetal development. Dev. Cell 51, 357–373 (2019).
Konnikova, L. et al. High-dimensional immune phenotyping and transcriptional analyses reveal robust recovery of viable human immune and epithelial cells from frozen gastrointestinal tissue. Mucosal Immunol. 11, 1684–1693 (2018).
Xin, H. et al. GMM-Demux: sample demultiplexing, multiplet detection, experiment planning, and novel cell-type verification in single cell sequencing. Genome Biol. 21, 188 (2020).
Luo, J., Erb, C. A. & Chen, K. Simultaneous measurement of surface proteins and gene expression from single cells. In: T-Cell Receptor Signaling Vol. 2111 35–46 (ed Liu, C.) (Springer, 2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Van den Brink, S. C. et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995).
Van de Sande, B. et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020).
Massalha, H. et al. A single cell atlas of the human liver tumor microenvironment. Mol. Syst. Biol. 16, e9682 (2020).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Scharfmann, R., Staels, W. & Albagli, O. The supply chain of human pancreatic β cell lines. J. Clin. Invest. 129, 3511–3520 (2019).
Sender, R. & Milo, R. The distribution of cellular turnover in the human body. Nat. Med. 27, 45–48 (2021).
Salomon, L. J., Bernard, J. P. & Ville, Y. Estimation of fetal weight: reference range at 20–36 weeks’ gestation and comparison with actual birth-weight reference range. Ultrasound Obstet. Gynecol. 29, 550–555 (2007).
Manco, R. et al. Clump sequencing exposes the spatial expression programs of intestinal secretory cells. Nat. Commun. 12, 3074 (2021).
Buse, M. G., Roberts, W. J. & Buse, J. The role of the human placenta in the transfer and metabolism of insulin. J. Clin. Invest. 41, 29–41 (1962).
Olaloye, O. O. et al. CD16+CD163+ monocytes traffic to sites of inflammation during necrotizing enterocolitis in premature infants. J. Exp. Med. 218, e20200344 (2021).
Acknowledgements
We thank Y. Dor and R. Scharfmann for insightful comments. S.I. is supported by the Wolfson Family Charitable Trust, the Edmond de Rothschild Foundations, the Fannie Sherr Fund, the Dr. Beth Rom-Rymer Stem Cell Research Fund, the Minerva Stiftung grant, Israel Science Foundation grant number 1486/16, Broad Institute‐Israel Science Foundation grant number 2615/18, the European Research Council under the European Union’s Horizon 2020 Research and Innovation Program (grant no. 768956), Chan Zuckerberg Initiative grant number CZF2019‐002434, the network of Pancreatic Organ Donors with Diabetes (nPOD), the Bert L. and N. Kuggie Vallee Foundation and the Howard Hughes Medical Institute international research scholar award. L.K. is supported by previous start-up funds from the University of Pittsburgh and current start-up funds from Yale University, Binational Science Foundation award number 2019075 and National Institute of Health (NIH) grants R21TR002639 and R21HD102565. No NIH funds were used for the fetal work of these studies.
Author information
Authors and Affiliations
Contributions
S.I., L.K. and A.E. conceived the study. D.L. and B.M. were involved in sample collection, processing and preparation for single-cell analysis. X.A., F.W. and K.C. were involved in library preparation. A.E. performed computational analysis, smFISH + immunofluorescence experiments and image analysis. L.F. designed the combined protocol. L.F. and K.B.H. performed smFISH + immunofluorescence experiments. L.K. and S.I. supervised the entirety of the project. All authors approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Medicine thanks Dominic Grun, Gordon Weir and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Anna Maria Ranzoni was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analysis of differentially-expressed genes between the human fetal and neonatal small intestines.
a-b, UMAP of small intestinal cells colored by age (a) or subject (b). c, MA plot showing differentially expressed genes between fetal and neonatal cells stratified by cell type. Genes in red have q-values below 0.1 and expression above 5e-5. INS, the most up-regulated gene in fetal enteroendocrine cells, is highlighted with a blue box.
Extended Data Fig. 2 Characterization of the fetal enteroendocrine cell types.
a, MA plot showing differentially expressed genes between fetal and neonatal enteroendocrine cells stratified by cell type. Genes in red have q-values below 0.05 and expression above 1e-4. INS, the most up-regulated gene in fetal K/L cells, is highlighted with a blue box. b, Spearman correlation distances between adult beta cells and the fetal endocrine cell types. Each dot represents a distance obtained from one of 100 bootstrap iterations, in which cells were sampled with replacement from the complete dataset. Distances from beta cells are significantly smaller for FIKL cells compared to all other enteroendocrine cell types (two-sided Wilcoxon rank-sum p = 1e-31 for the differences between FIKL cells and K/L INS- cells, the second closest enteroendocrine cell population). White circles are medians, gray boxes mark the 25–75 percentiles, gray lines extend from 1.5 times the interquartile range (IQR) above the 75 percentile to 1.5 times the IQR below the 25 percentile and truncated at the minimal or maximal measured values. c, RGS16 expression (log10 of the sum-normalized UMI counts).
Extended Data Fig. 3 Analysis of gene regulatory networks.
a, Clustergram of TF activities Z-score of mean AUC for each cell type. The list includes TFs with the most differential activities in the different cell types (Methods). b-g, Selected TFs from (a). In b-g, each dot is a cell, white circles are medians, gray boxes mark the 25–75 percentiles, gray lines extend from 1.5 times the interquartile range (IQR) above the 75 percentile to 1.5 times the IQR below the 25 percentile and truncated at the minimal or maximal measured values. Black lines connect the means.
Extended Data Fig. 4 FIKL cells appear in earlier developmental time points.
a-b, Combined UMAP of 1st trimester14 and 2nd trimester colored by enteroendocrine subtype (a) or INS expression (log10(normalized expression)), panel b. Dots with a black outline in a-b are 1st trimester cells, dots without an outline are 2nd trimester cells. c, Heatmap of INS + cells enrichment in each enteroendocrine subtype. Colors are –log10(hypergeometric p-value for the enrichment of INS + cells within each cell cluster).
Extended Data Fig. 5 In-situ validation of FIKL cells at different developmental stages.
a, Intestinal section with a typical example of an INS + cell marked with INS protein (green) from a 12 weeks GA fetus. Scale bar - 50 µm. b, Blowup of the INS + cell from (a) stained with both INS-mRNA (magenta) and INS-protein (green). Scale bar - 5 µm. c-f, Representatives examples of INS + cells in small intestine tissues from 12 weeks GA (c, 3 cells out of 20 found), 14 weeks GA (d, 2 cells out of 3 found), 18 weeks GA (e, 1 cell out of 1 found) and 21 weeks GA (f, 3 cells out of 23 found) stained with INS-mRNA (magenta), INS-protein (green). Scale bar - 5 µm. In all images nuclei are stained by DAPI (blue).
Extended Data Fig. 6 Insulin is not detected in embryonic mouse small intestine.
a, Mouse embryo intestinal section (E16.5) stained with the pan-enteroendocrine cell marker Chga (gray) and blowups of selected Chga+ cells. Scalebar - 50 µm for big image and 10 µm for blowups. b, Mouse embryo pancreatic tissue stained with Ins2-mRNA (magenta) and INS-protein (green) shown as a positive control for the in-situ detection of mouse cells expressing insulin. Scale bar - 10 µm. In all images nuclei are stained by DAPI (blue). Images are representative of 6 E16.5 embryonic subjects and 5 P0 neonatal subjects.
Supplementary information
Supplementary Tables
Supplementary Tables 1–9
Rights and permissions
About this article
Cite this article
Egozi, A., Llivichuzhca-Loja, D., McCourt, B.T. et al. Insulin is expressed by enteroendocrine cells during human fetal development. Nat Med 27, 2104–2107 (2021). https://doi.org/10.1038/s41591-021-01586-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-021-01586-1
This article is cited by
-
Single-nucleus multi-omics of human stem cell-derived islets identifies deficiencies in lineage specification
Nature Cell Biology (2023)
-
A guide from the stomach to β cells
Nature Cell Biology (2023)
-
Stomach-derived human insulin-secreting organoids restore glucose homeostasis
Nature Cell Biology (2023)
-
Anatomical and functional maturation of the mid-gestation human enteric nervous system
Nature Communications (2023)