Abstract
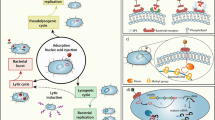
Although bacteriophages have been overshadowed as therapeutic agents by antibiotics for decades, the emergence of multidrug-resistant bacteria and a better understanding of the role of the gut microbiota in human health and disease have brought them back into focus. In this Perspective, we briefly introduce basic phage biology and summarize recent discoveries about phages in relation to their role in the gut microbiota and gastrointestinal diseases, such as inflammatory bowel disease and chronic liver disease. In addition, we review preclinical studies and clinical trials of phage therapy for enteric disease and explore current challenges and potential future directions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ni, J., Wu, G. D., Albenberg, L. & Tomov, V. T. Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 14, 573–584 (2017).
Tripathi, A. et al. The gut–liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411 (2018).
Wong, S. H. & Yu, J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 16, 690–704 (2019).
Young, V. B. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ 356, j831 (2017).
Lurie-Weinberger, M. N. & Gophna, U. Archaea in and on the human body: health implications and future directions. PLOS Pathog. 11, e1004833 (2015).
Chu, H. et al. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J. Hepatol. 72, 391–400 (2020).
Lang, S. et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology 71, 522–538 (2020).
Norman, J. M. et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 (2015).
Clooney, A. G. et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26, 764–778 (2019).
Zuo, T. et al. Gut mucosal virome alterations in ulcerative colitis. Gut 68, 1169–1179 (2019).
Hannigan, G. D., Duhaime, M. B., Ruffin, M. T., Koumpouras, C. C. & Schloss, P. D. Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio 9, e02248-18 (2018).
Nakatsu, G. et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 155, 529–541 (2018).
Jiang, L. et al. Intestinal virome in patients with alcoholic hepatitis. Hepatology 72, 2182–2196 (2020).
Lang, S. et al. Intestinal virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology 159, 1839–1852 (2020).
Shkoporov, A. N. & Hill, C. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25, 195–209 (2019).
Twort, F. W. An investigation on the nature of ultra-microscopic viruses. Lancet 186, 1241–1243 (1915).
d’Herelle, F. Sur un microbe invisible antagoniste des bacilles dysenteriques [French]. C. R. Acad. Sci. 165, 373–375 (1917).
Summers, W. C. Bacteriophage therapy. Annu. Rev. Microbiol. 55, 437–451 (2001).
Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenza. Br. J. Exp. Pathol. 10, 226–236 (1929).
Aminov, R. History of antimicrobial drug discovery: major classes and health impact. Biochem. Pharmacol. 133, 4–19 (2017).
Goossens, H., Ferech, M., Stichele, R. V. & Elseviers, M. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365, 579–587 (2005).
Spellberg, B., Bartlett, J. G. & Gilbert, D. N. The future of antibiotics and resistance. N. Engl. J. Med. 368, 299–302 (2013).
Becattini, S., Taur, Y. & Pamer, E. G. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol. Med. 22, 458–478 (2016).
Vila, A. V. et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 11, 362 (2020).
Bobay, L. M. & Ochman, H. Biological species in the viral world. Proc. Natl Acad. Sci. USA 115, 6040–6045 (2018).
Mushegian, A. R. Are there 1031 virus particles on earth, or more, or fewer? J. Bacteriol. 202, e00052-20 (2020).
Rohwer, F. Global phage diversity. Cell 113, 141 (2003).
Clokie, M. R. J., Millard, A. D., Letarov, A. V. & Heaphy, S. Phages in nature. Bacteriophage 1, 31–45 (2011).
Ashelford, K. E., Day, M. J. & Fry, J. C. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69, 285–289 (2003).
Pratama, A. A. & van Elsas, J. D. The ‘neglected’ soil virome–potential role and impact. Trends Microbiol. 26, 649–662 (2018).
Suttle, C. A. Viruses in the sea. Nature 437, 356–361 (2005).
Suttle, C. A. Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007).
Paez-Espino, D. et al. Uncovering earth’s virome. Nature 536, 425–430 (2016).
Manrique, P. et al. Healthy human gut phageome. Proc. Natl Acad. Sci. USA 113, 10400–10405 (2016).
Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010).
Reyes, A., Semenkovich, N. P., Whiteson, K., Rohwer, F. & Gordon, J. I. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 10, 607–617 (2012).
Hendrix, R. W. Bacteriophage genomics. Curr. Opin. Microbiol. 6, 506–511 (2003).
Hatfull, G. F. Bacteriophage genomics. Curr. Opin. Microbiol. 11, 447–453 (2008).
Ackermann, H. W. & Prangishvili, D. Prokaryote viruses studied by electron microscopy. Arch. Virol. 157, 1843–1849 (2012).
Dion, M. B., Oechslin, F. & Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 18, 125–138 (2020).
Koonin, E. V. et al. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 84, e00061-19 (2020).
Al-Shayeb, B. et al. Clades of huge phages from across earth’s ecosystems. Nature 578, 425–431 (2020).
Ackermann, H. W. in The Bacteriophages 2nd edn (ed. Calendar, R. L.) 8–16 (Oxford Univ. Press, 2006).
Goldberg, E. B. in Bacteriophage T4 (eds Mathews, C. K., Kutter, E. M., Mosig, G. & Berget, P. B.) 32–39 (American Society for Microbiology, 1983).
Shkoporov, A. N. et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26, 527–541 (2019).
Gregory, A. C. et al. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe 28, 724–740 (2020).
Goldberg, E. B. in Receptors and Recognition (Series B, Volume 7): Virus Receptors (Part 1: Bacterial Viruses) (eds Yamamura, H. I. & Enna, S. J.) 115–141 (Chapman & Hall, 1981).
Campbell, A. The future of bacteriophage biology. Nat. Rev. Genet. 4, 471–477 (2003).
Ofir, G. & Sorek, R. Contemporary phage biology: from classic models to new insights. Cell 172, 1260–1270 (2018).
Lindberg, A. A. Bacteriophage receptors. Annu. Rev. Microbiol. 27, 205–241 (1973).
Ge, H. et al. The “fighting wisdom and bravery” of tailed phage and host in the process of adsorption. Microbiol. Res. 230, 126344 (2020).
Silva, J. B., Storms, Z. & Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 363, fnw002 (2016).
Young, R. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56, 430–481 (1992).
Cahill, J. & Young, R. in Advances in Virus Research Vol. 103 Ch. 2 (eds Kielian, M., Mettenleiter, T. C. & Roosinck, M. J.) 33–70 (Academic Press, 2019).
Zeng, L. et al. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell 141, 682–691 (2010).
Dou, C. et al. Structural and functional insights into the regulation of the lysis–lysogeny decision in viral communities. Nat. Microbiol. 3, 1285–1294 (2018).
Feiner, R. et al. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 13, 641–650 (2015).
Howard-Varona, C., Hargreaves, K. R., Abedon, S. T. & Sullivan, M. B. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11, 1511–1520 (2017).
Bondy-Denomy, J. et al. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 10, 2854–2866 (2016).
Fortier, L. C. & Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4, 354–365 (2013).
Banks, D. J., Lei, B. & Musser, J. M. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 71, 7079–7086 (2003).
Goerke, C., Köller, J. & Wolz, C. Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob. Agents Chemother. 50, 171–177 (2006).
Choi, J., Kotay, S. M. & Goel, R. Various physico-chemical stress factors cause prophage induction in Nitrosospira multiformis 25196–an ammonia oxidizing bacteria. Water Res. 44, 4550–4558 (2010).
Alexeeva, S., Guerra Martínez, J. A., Spus, M. & Smid, E. J. Spontaneously induced prophages are abundant in a naturally evolved bacterial starter culture and deliver competitive advantage to the host. BMC Microbiol. 18, 120 (2018).
Nanda, A. M., Thormann, K. & Frunzke, J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host–microbe interactions. J. Bacteriol. 197, 410–419 (2015).
Labrie, S. J., Samson, J. E. & Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010).
Samson, J. E., Magadán, A. H., Sabri, M. & Moineau, S. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687 (2013).
Ackermann, H. W. & Dubow, M. S. Viruses of Prokaryotes Vol. 1: General Properties of Bacteriophages 49–85 (CRC Press, 1987).
Dufour, N. et al. Bacteriophage LM33_P1, a fast-acting weapon against the pandemic ST131-O25b:H4 Escherichia coli clonal complex. J. Antimicrob. Chemother. 71, 3072–3080 (2016).
Kaiser, D. & Dworkin, M. Gene transfer to myxobacterium by Escherichia coli phage P1. Science 187, 653–654 (1975).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Huttenhower, C. et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Lloyd-Price, J. et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Lourenço, M. et al. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28, 390–401 (2020).
Sender, R., Fuchs, S. & Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340 (2016).
Breitbart, M. et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 (2003).
Kim, M. S., Park, E. J., Roh, S. W. & Bae, J. W. Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 77, 8062–8070 (2011).
Liang, G. et al. The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature 581, 470–474 (2020).
Reyes, A. et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl Acad. Sci. USA 112, 11941–11946 (2015).
Lim, E. S. et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234 (2015).
Lim, E. S., Wang, D. & Holtz, L. R. The bacterial microbiome and virome milestones of infant development. Trends Microbiol. 24, 801–810 (2016).
Edwards, R. A. et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 4, 1727–1736 (2019).
Moreno-Gallego, J. L. et al. Virome diversity correlates with intestinal microbiome diversity in adult monozygotic twins. Cell Host Microbe 25, 261–272 (2019).
Tap, J. et al. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11, 2574–2584 (2009).
Pedulla, M. L. et al. Origins of highly mosaic mycobacteriophage genomes. Cell 113, 171–182 (2003).
Deng, L. et al. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513, 242–245 (2014).
Mavrich, T. N. & Hatfull, G. F. Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol. 2, 17112 (2017).
Hendrix, R. W., Smith, M. C. M., Burns, R. N., Ford, M. E. & Hatfull, G. F. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc. Natl Acad. Sci. USA 96, 2192–2197 (1999).
Shapiro, J. W. & Putonti, C. Gene co-occurrence networks reflect bacteriophage ecology and evolution. mBio 9, e01870-17 (2018).
Shkoporov, A. N. et al. Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome 6, 68 (2018).
Conceição-Neto, N. et al. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci. Rep. 5, 16532 (2015).
Waller, A. S. et al. Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J. 8, 1391–1402 (2014).
Ma, Y., You, X., Mai, G., Tokuyasu, T. & Liu, C. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 6, 24 (2018).
Kleiner, M., Hooper, L. V. & Duerkop, B. A. Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genomics 16, 7 (2015).
Flewett, T. H., Bryden, A. S. & Davies, H. Diagnostic electron microscopy of faeces. J. Clin. Pathol. 27, 603–608 (1974).
Kim, K. H. & Bae, J. W. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl. Environ. Microbiol. 77, 7663–7668 (2011).
Yilmaz, S., Allgaier, M. & Hugenholtz, P. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat. Methods 7, 943–944 (2010).
Roux, S. et al. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ 4, e2777 (2016).
Krishnamurthy, S. R. & Wang, D. Origins and challenges of viral dark matter. Virus Res. 239, 136–142 (2017).
Santiago-Rodriguez, T. M. & Hollister, E. B. Human virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses 11, 656 (2019).
Barr, J. J. et al. Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc. Natl Acad. Sci. USA 110, 10771–10776 (2013).
Wagner, J. et al. Bacteriophages in gut samples from pediatric Crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm. Bowel Dis. 19, 1598–1608 (2013).
Siringan, P., Connerton, P. L., Cummings, N. J. & Connerton, I. F. Alternative bacteriophage life cycles: the carrier state of Campylobacter jejuni. Open. Biol. 4, 130200 (2014).
Pourcel, C., Midoux, C., Vergnaud, G. & Latino, L. A carrier state is established in Pseudomonas aeruginosa by phage LeviOr01, a newly isolated ssRNA levivirus. J. Gen. Virol. 98, 2181–2189 (2017).
Joossens, M. et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60, 631–637 (2011).
Lax, S. et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 (2014).
Zmora, N., Suez, J. & Elinav, E. You are what you eat: diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 16, 35–56 (2019).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Zuo, T. et al. Human-gut-DNA virome variations across geography, ethnicity, and urbanization. Cell Host Microbe 28, 741–751 (2020).
Coughlan, S. et al. The gut virome in irritable bowel syndrome differs from that of controls. Gut Microbes 13, 1–15 (2021).
Lepage, P. et al. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 57, 424–425 (2008).
Bajaj, J. S. et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut 70, 1162–1173 (2021).
d’Herelle, F. & Smith, G. H. The bacteriophage and its behavior 490–497 (Williams & Wilkins, 1926).
Spence, R. C. & Mckinley, E. B. The therapeutic value of the bacteriophage in treatment of bacillary dysentery. South. Med. J. 17, 563–571 (1924).
Burnet, F. M., McKie, M. & Wood, I. J. Investigations on bacillary dysentery in infants, with special reference to bacteriophage phenomena. Med. J. Aust. 2, 71–78 (1930).
D’Herelle, F. Studies upon Asiatic cholera. Yale J. Biol. Med. 1, 195–219 (1929).
Becherich, A. & Hauduroy, P. Le bactériophage dans le traitement de la fièvre typhoïde [French]. C. R. Soc. Biol. 86, 168–170 (1922).
Smith, J. The bacteriophage in the treatment of typhoid fever. Br. Med. J. 2, 47–49 (1924).
Hadley, P. The Twort-D’Herelle phenomenon: a critical review and presentation of a new conception (homogamic theory) of bacteriophage action. J. Infect. Dis. 42, 263–434 (1928).
Eaton, M. D. & Bayne-Jones, S. Bacteriophage therapy: review of the principles and results of the use of bacteriophage in the treatment of infections. J. Am. Med. Assoc. 103, 1769–1776 (1934).
Merabishvili, M. et al. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE 4, e4944 (2009).
Bruttin, A. & Brüssow, H. Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob. Agents Chemother. 49, 2874–2878 (2005).
Sarker, S. A. et al. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 434, 222–232 (2012).
McCallin, S. et al. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology 443, 187–196 (2013).
Sarker, S. A. et al. Oral application of Escherichia coli bacteriophage: safety tests in healthy and diarrheal children from Bangladesh. Environ. Microbiol. 19, 237–250 (2017).
Sarker, S. A. et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine 4, 124–137 (2016).
Schooley, R. T. et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61, e00954-17 (2017).
Jennes, S. et al. Use of bacteriophages in the treatment of colistin-only-sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury–a case report. Crit. Care 21, 129 (2017).
Dedrick, R. M. et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733 (2019).
Yen, M., Cairns, L. S. & Camilli, A. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat. Commun. 8, 14187 (2017).
Faruque, S. M. et al. Self-limiting nature of seasonal cholera epidemics: role of host-mediated amplification of phage. Proc. Natl Acad. Sci. USA 102, 6119–6124 (2005).
Khoruts, A. & Sadowsky, M. J. Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 13, 508–516 (2016).
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R. & Rastall, R. A. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616 (2019).
Deaton, J., Ertle, E. & Dawson, H. G. Prebiotic compositions comprising one or more types of bacteriophage. US Patent 9,839,657 (2017).
Gindin, M., Febvre, H. P., Rao, S., Wallace, T. C. & Weir, T. L. Bacteriophage for Gastrointestinal Health (PHAGE) study: evaluating the safety and tolerability of supplemental bacteriophage consumption. J. Am. Coll. Nutr. 38, 68–75 (2019).
Grubb, D. S. et al. PHAGE-2 study: supplemental bacteriophages extend Bifidobacterium animalis subsp. lactis BL04 benefits on gut health and microbiota in healthy adults. Nutrients 12, 2474 (2020).
Rolhion, N. & Darfeuille-Michaud, A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm. Bowel Dis. 13, 1277–1283 (2007).
Palmela, C. et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 67, 574–587 (2018).
Galtier, M. et al. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J. Crohn’s Colitis 11, 840–847 (2017).
Duan, Y. et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511 (2019).
McCoy, W. C. & Mason, J. M. 3rd Enterococcal endocarditis associated with carcinoma of the sigmoid; report of a case. J. Med. Assoc. State Ala. 21, 162–166 (1951).
Abdulamir, A. S., Hafidh, R. R. & Bakar, F. A. The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its etiological role. J. Exp. Clin. Cancer Res. 30, 11 (2011).
Boleij, A., van Gelder, M. M. H. J., Swinkels, D. W. & Tjalsma, H. Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin. Infect. Dis. 53, 870–878 (2011).
Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689 (2019).
Yachida, S. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25, 968–976 (2019).
Wong, S. H. et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 66, 1441–1448 (2017).
Castellarin, M. et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012).
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013).
Bullman, S. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017).
Wu, S. et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022 (2009).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Dejea, C. M. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018).
Huycke, M. M., Abrams, V. & Moore, D. R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23, 529–536 (2002).
Wang, X. et al. 4-Hydroxy-2-nonenal mediates genotoxicity and bystander effects caused by Enterococcus faecalis–infected macrophages. Gastroenterology 142, 543–551 (2012).
Kumar, R. et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog. 13, e1006440 (2017).
Zhu, W. et al. Editing of the gut microbiota reduces carcinogenesis in mouse models of colitis-associated colorectal cancer. J. Exp. Med. 216, 2378–2393 (2019).
Henning, U. et al. in Molecular Biology of Bacteriophage T4 Vol. 1 Ch. 23 (ed. Karam, J. D.) 291–298 (ASM Press, 1994).
Iida, S. Bacteriophage P1 carries two related sets of genes determining its host range in the invertible C segment of its genome. Virology 134, 421–434 (1984).
Summer, E. J. et al. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. J. Mol. Biol. 340, 49–65 (2004).
Liu, M. et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295, 2091–2094 (2002).
Bar, H., Yacoby, I. & Benhar, I. Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC Biotechnol. 8, 37 (2008).
Yacoby, I., Shamis, M., Bar, H., Shabat, D. & Benhar, I. Targeting antibacterial agents by using drug-carrying filamentous bacteriophages. Antimicrob. Agents Chemother. 50, 2087–2097 (2006).
Yacoby, I., Bar, H. & Benhar, I. Targeted drug-carrying bacteriophages as antibacterial nanomedicines. Antimicrob. Agents Chemother. 51, 2156–2163 (2007).
Vaks, L. & Benhar, I. in Biomedical Nanotechnology: Methods in Molecular Biology (Methods and Protocols) Vol. 726 (ed. Hurst, S.) 187–206 (Humana Press, 2011).
Zheng, D. W. et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 3, 717–728 (2019).
Yu, T. C. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563 (2017).
Dong, X. et al. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci. Adv. 6, eaba1590 (2020).
Devkota, S. et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–108 (2012).
Hendrikx, T. et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68, 1504–1515 (2019).
Singh, N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 (2014).
Cani, P. D. & Jordan, B. F. Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 15, 671–682 (2018).
Febvre, H. P. et al. PHAGE study: effects of supplemental bacteriophage intake on inflammation and gut microbiota in healthy adults. Nutrients 11, 666 (2019).
Sweere, J. M. et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 363, eaat9691 (2019).
Tetz, G. V. et al. Bacteriophages as potential new mammalian pathogens. Sci. Rep. 7, 7043 (2017).
Van Belleghem, J. D., Clement, F., Merabishvili, M., Lavigne, R. & Vaneechoutte, M. Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci. Rep. 7, 8004 (2017).
Gogokhia, L. et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe 25, 285–299 (2019).
Zhang, L. et al. Staphylococcus aureus bacteriophage suppresses LPS-induced inflammation in MAC-T bovine mammary epithelial cells. Front. Microbiol. 9, 1614 (2018).
Hong, Y. et al. The impact of orally administered phages on host immune response and surrounding microbial communities. Bacteriophage 6, e1211066 (2016).
Miernikiewicz, P. et al. T4 phage and its head surface proteins do not stimulate inflammatory mediator production. PLoS ONE 8, e71036 (2013).
Merabishvili, M., Pirnay, J. P. & De Vos, D. in Bacteriophage Therapy: From Lab to Clinical Practice (eds Azeredo, J. & Sillankorva, S.) 99–110 (Springer, 2018).
Betts, A., Vasse, M., Kaltz, O. & Hochberg, M. E. Back to the future: evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas aeruginosa PAO1. Evolut. Appl. 6, 1054–1063 (2013).
Friman, V. P. et al. Pre-adapting parasitic phages to a pathogen leads to increased pathogen clearance and lowered resistance evolution with Pseudomonas aeruginosa cystic fibrosis bacterial isolates. J. Evolut. Biol. 29, 188–198 (2016).
Dunne, M. et al. Reprogramming bacteriophage host range through structure-guided design of chimeric receptor binding proteins. Cell Rep. 29, 1336–1350 (2019).
Yehl, K. et al. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell 179, 459–469 (2019).
Ly-Chatain, M. H. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 5, 51 (2014).
Cui, Z., Guo, X., Feng, T. & Li, L. Exploring the whole standard operating procedure for phage therapy in clinical practice. J. Transl. Med. 17, 373 (2019).
Fabijan, A. P. et al. Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat. Microbiol. 5, 465–472 (2020).
Jończyk, E., Kłak, M., Międzybrodzki, R. & Górski, A. The influence of external factors on bacteriophages–review. Folia Microbiol. 56, 191–200 (2011).
Colom, J. et al. Microencapsulation with alginate/CaCO3: a strategy for improved phage therapy. Sci. Rep. 7, 41441 (2017).
Hsu, B. B. et al. In situ reprogramming of gut bacteria by oral delivery. Nat. Commun. 11, 5030 (2020).
Dąbrowska, K. Phage therapy: what factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Med. Res. Rev. 39, 2000–2025 (2019).
Srivastava, A. S., Kaido, T. & Carrier, E. Immunological factors that affect the in vivo fate of T7 phage in the mouse. J. Virol. Meth. 115, 99–104 (2004).
Merril, C. R. et al. Long-circulating bacteriophage as antibacterial agents. Proc. Natl Acad. Sci. USA 93, 3188–3192 (1996).
Capparelli, R., Parlato, M., Borriello, G., Salvatore, P. & Iannelli, D. Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob. Agents Chemother. 51, 2765–2773 (2007).
Capparelli, R., Ventimiglia, I., Roperto, S., Fenizia, D. & Iannelli, D. Selection of an Escherichia coli O157:H7 bacteriophage for persistence in the circulatory system of mice infected experimentally. Clin. Microbiol. Infect. 12, 248–253 (2006).
Fauconnier, A. in Bacteriophage Therapy: From Lab to Clinical Practice (eds Azeredo, J. & Sillankorva, S.) 253–268 (Springer, 2018).
Pirnay, J. P. et al. The phage therapy paradigm: prêt-à-porter or sur-mesure? Pharm. Res. 28, 934–937 (2011).
Servick, K. Beleaguered phage therapy trial presses on. Science 352, 1506 (2016).
Pirnay, J. P. et al. The magistral phage. Viruses 10, 64 (2018).
EUR-Lex. Opinion of Advocate General Szpunar delivered on 30 June 2016. Case C-276/15: Hecht-Pharma GmbH v Hohenzollern Apotheke, owned by Winfried Ertelt. EUR-Lex https://eur-lex.europa.eu/legal-content/en/TXT/?uri=CELEX:62015CC0276 (2016).
Corbellino, M. et al. Eradication of a multidrug-resistant, carbapenemase-producing Klebsiella pneumoniae isolate following oral and intra-rectal therapy with a custom made, lytic bacteriophages preparation. Clin. Infect. Dis. 70, 1998–2001 (2020).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT03808103 (2021).
Acknowledgements
B.S. was supported in part by a Biocodex Microbiota Foundation Grant, NIH grants R01 AA024726, U01 AA026939, by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and services provided by P30 DK120515 and P50 AA011999.
Author information
Authors and Affiliations
Contributions
Y.D. researched data for the article, made a substantial contribution to discussion of content, wrote the article, and reviewed/edited the manuscript before submission. R.Y. and B.S. made a substantial contribution to discussion of content and reviewed/edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
B.S. has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics, Mabwell Therapeutics, Patara Pharmaceuticals and Takeda. B.S.’s institution UC San Diego has received grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, Synlogic Operating Company, Prodigy Biotech and Axial Biotherapeutics. B.S. is founder of Nterica Bio. UC San Diego has filed several patents with B.S. and Y.D. as inventors related to this work. R.Y. was formerly involved with GangaGen (Bangalore, India) as a member of its scientific advisory board.
Additional information
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks Harald Brüssow, Laurent Debarbieux and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Duan, Y., Young, R. & Schnabl, B. Bacteriophages and their potential for treatment of gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 19, 135–144 (2022). https://doi.org/10.1038/s41575-021-00536-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-021-00536-z
This article is cited by
-
Therapeutics for neurodegenerative diseases by targeting the gut microbiome: from bench to bedside
Translational Neurodegeneration (2024)
-
Identification of HDV-like theta ribozymes involved in tRNA-based recoding of gut bacteriophages
Nature Communications (2024)
-
Characterizations of the multi-kingdom gut microbiota in Chinese patients with gouty arthritis
BMC Microbiology (2023)
-
A balanced gut microbiota is essential to maintain health in captive sika deer
Applied Microbiology and Biotechnology (2022)