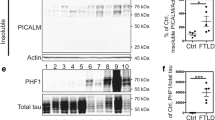
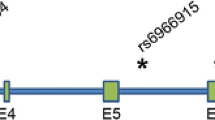
Abstract
Primary age-related tauopathy (PART) is a neurodegenerative pathology with features distinct from but also overlapping with Alzheimer disease (AD). While both exhibit Alzheimer-type temporal lobe neurofibrillary degeneration alongside amnestic cognitive impairment, PART develops independently of amyloid-β (Aβ) plaques. The pathogenesis of PART is not known, but evidence suggests an association with genes that promote tau pathology and others that protect from Aβ toxicity. Here, we performed a genetic association study in an autopsy cohort of individuals with PART (n = 647) using Braak neurofibrillary tangle stage as a quantitative trait. We found some significant associations with candidate loci associated with AD (SLC24A4, MS4A6A, HS3ST1) and progressive supranuclear palsy (MAPT and EIF2AK3). Genome-wide association analysis revealed a novel significant association with a single nucleotide polymorphism on chromosome 4 (rs56405341) in a locus containing three genes, including JADE1 which was significantly upregulated in tangle-bearing neurons by single-soma RNA-seq. Immunohistochemical studies using antisera targeting JADE1 protein revealed localization within tau aggregates in autopsy brains with four microtubule-binding domain repeats (4R) isoforms and mixed 3R/4R, but not with 3R exclusively. Co-immunoprecipitation in post-mortem human PART brain tissue revealed a specific binding of JADE1 protein to four repeat tau lacking N-terminal inserts (0N4R). Finally, knockdown of the Drosophila JADE1 homolog rhinoceros (rno) enhanced tau-induced toxicity and apoptosis in vivo in a humanized 0N4R mutant tau knock-in model, as quantified by rough eye phenotype and terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) in the fly brain. Together, these findings indicate that PART has a genetic architecture that partially overlaps with AD and other tauopathies and suggests a novel role for JADE1 as a modifier of neurofibrillary degeneration.





Similar content being viewed by others
References
Abner EL, Neltner JH, Jicha GA, Patel E, Anderson SL, Wilcock DM et al (2018) Diffuse amyloid-beta plaques, neurofibrillary tangles, and the impact of APOE in elderly persons’ brains lacking neuritic amyloid plaques. J Alzh Dis 64:1307–1324. https://doi.org/10.3233/Jad-180514
Aldridge GM, Podrebarac DM, Greenough WT, Weiler IJ (2008) The use of total protein stains as loading controls: an alternative to high-abundance single-protein controls in semi-quantitative immunoblotting. J Neurosci Methods 172:250–254. https://doi.org/10.1016/j.jneumeth.2008.05.003
Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT (2010) Data quality control in genetic case-control association studies. Nat Protoc 5:1564–1573. https://doi.org/10.1038/nprot.2010.116
Andrews SJ, Fulton-Howard B, Goate A (2020) Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol 19:326–335. https://doi.org/10.1016/S1474-4422(19)30435-1
Augustinack JC, Schneider A, Mandelkow EM, Hyman BT (2002) Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer’s disease. Acta Neuropathol 103:26–35. https://doi.org/10.1007/s004010100423
Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ et al (2014) Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer’s disease and related dementias. Plos Genet. https://doi.org/10.1371/journal.pgen.1004606
Bell WR, An Y, Kageyama Y, English C, Rudow GL, Pletnikova O et al (2019) Neuropathologic, genetic, and longitudinal cognitive profiles in primary age-related tauopathy (PART) and Alzheimer’s disease. Alzheimers Dement 15:8–16. https://doi.org/10.1016/j.jalz.2018.07.215
Bennett DA, Schneider JA, Arvanitakis Z, Kelly JF, Aggarwal NT, Shah RC et al (2006) Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology 66:1837–1844. https://doi.org/10.1212/01.wnl.0000219668.47116.e6
Besser LM, Crary JF, Mock C, Kukull WA (2017) Comparison of symptomatic and asymptomatic persons with primary age-related tauopathy. Neurology 89:1707–1715. https://doi.org/10.1212/WNL.0000000000004521
Besser LM, Mock C, Teylan MA, Hassenstab J, Kukull WA, Crary JF (2019) Differences in cognitive impairment in primary age-related tauopathy versus alzheimer disease. J Neuropath Exp Neur 78:219–228. https://doi.org/10.1093/jnen/nly132
Boettger LM, Handsaker RE, Zody MC, McCarroll SA (2012) Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet 44:881–882. https://doi.org/10.1038/ng.2334
Boone DK, Weisz HA, Bi M, Falduto MT, Torres KEO, Willey HE et al (2017) Evidence linking microRNA suppression of essential prosurvival genes with hippocampal cell death after traumatic brain injury. Sci Rep 7:6645. https://doi.org/10.1038/s41598-017-06341-6
Borgal L, Habbig S, Hatzold J, Liebau MC, Dafinger C, Sacarea I et al (2012) The ciliary protein nephrocystin-4 translocates the canonical Wnt regulator Jade-1 to the nucleus to negatively regulate beta-catenin signaling. J Biol Chem 287:25370–25380. https://doi.org/10.1074/jbc.M112.385658
Braak E, Braak H, Mandelkow EM (1994) A sequence of cytoskeleton changes related to the formation of neurofibrillary tangles and neuropil threads. Acta Neuropathol 87:554–567. https://doi.org/10.1007/BF00293315
Braak H, Braak E (1995) Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging 16:271–278. https://doi.org/10.1016/0197-4580(95)00021-6
Braak H, Del Tredici K (2014) Are cases with tau pathology occurring in the absence of A beta deposits part of the AD-related pathological process? Acta Neuropathol 128:767–772. https://doi.org/10.1007/s00401-014-1356-1
Broce I, Karch CM, Wen N, Fan CC, Wang YP, Tan CH et al (2018) Immune-related genetic enrichment in frontotemporal dementia: an analysis of genome-wide association studies. Plos Med. https://doi.org/10.1371/journal.pmed.1002487
Chen JA, Chen Z, Won H, Huang AY, Lowe JK, Wojta K et al (2018) Joint genome-wide association study of progressive supranuclear palsy identifies novel susceptibility loci and genetic correlation to neurodegenerative diseases. Mol Neurodegener 13:41. https://doi.org/10.1186/s13024-018-0270-8
Chitalia VC, Foy RL, Bachschmid MM, Zeng L, Panchenko MV, Zhou MI et al (2008) Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol 10:1208–1216. https://doi.org/10.1038/ncb1781
Clarke L, Fairley S, Zheng-Bradley XQ, Streeter I, Perry E, Lowy E et al (2017) The international genome sample resource (IGSR): a worldwide collection of genome variation incorporating the 1000 genomes project data. Nucleic Acids Res 45:D854–D859. https://doi.org/10.1093/nar/gkw829
Combs B, Kanaan NM (2017) Exposure of the amino terminus of tau is a pathological event in multiple tauopathies. Am J Pathol 187:1222–1229. https://doi.org/10.1016/j.ajpath.2017.01.019
Congdon EE, Sigurdsson EM (2018) Tau-targeting therapies for Alzheimer disease. Nat Rev Neurol 14:399–415. https://doi.org/10.1038/s41582-018-0013-z
Crary JF, Trojanowski JQ, Schneider JA, Abisambra JF, Abner EL, Alafuzoff I et al (2014) Primary age-related tauopathy (PART): a common pathology associated with human aging. Acta Neuropathol 128:755–766. https://doi.org/10.1007/s00401-014-1349-0
Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A et al (2016) Next-generation genotype imputation service and methods. Nat Genet 48:1284–1287. https://doi.org/10.1038/ng.3656
De Jager PL, Ma Y, McCabe C, Xu J, Vardarajan BN, Felsky D et al (2018) A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci Data 5:180142. https://doi.org/10.1038/sdata.2018.142
Derisbourg M, Leghay C, Chiappetta G, Fernandez-Gomez FJ, Laurent C, Demeyer D et al (2015) Role of the Tau N-terminal region in microtubule stabilization revealed by new endogenous truncated forms. Sci Rep-Uk. https://doi.org/10.1038/srep09659
Dregni AJ, Mandala VS, Wu H, Elkins MR, Wang HK, Hung I et al (2019) In vitro 0N4R tau fibrils contain a monomorphic beta-sheet core enclosed by dynamically heterogeneous fuzzy coat segments. Proc Natl Acad Sci U S A 116:16357–16366. https://doi.org/10.1073/pnas.1906839116
Duyckaerts C, Braak H, Brion JP, Buee L, Del Tredici K, Goedert M et al (2015) PART is part of Alzheimer disease. Acta Neuropathol 129:749–756. https://doi.org/10.1007/s00401-015-1390-7
Efthymiou AG, Goate AM (2017) Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol Neurodegener 12:43. https://doi.org/10.1186/s13024-017-0184-x
Farfel JM, Yu L, De Jager PL, Schneider JA, Bennett DA (2016) Association of APOE with tau-tangle pathology with and without beta-amyloid. Neurobiol Aging 37:19–25. https://doi.org/10.1016/j.neurobiolaging.2015.09.011
Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK et al (2015) MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16:278. https://doi.org/10.1186/s13059-015-0844-5
Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ et al (2017) Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547:185–190. https://doi.org/10.1038/nature23002
Folstein MF, Robins LN, Helzer JE (1983) The mini-mental state examination. Arch Gen Psychiatry 40:812
Frost B, Ollesch J, Wille H, Diamond MI (2009) Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J Biol Chem 284:3546–3551. https://doi.org/10.1074/jbc.M805627200
Hoglinger GU, Melhem NM, Dickson DW, Sleiman PMA, Wang LS, Klei L et al (2011) Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet 43:699-U125. https://doi.org/10.1038/ng.859
Holmes BB, Devos SL, Kfoury N, Li M, Jacks R, Yanamandra K et al (2013) Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. P Natl Acad Sci USA 110:E3138–E3147. https://doi.org/10.1073/pnas.1301440110
Hong S, Prokopenko D, Dobricic V, Kilpert F, Bos I, Vos SJB et al (2020) Genome-wide association study of Alzheimer’s disease CSF biomarkers in the EMIF-AD multimodal biomarker discovery dataset. Trans Psychiatry 10:403. https://doi.org/10.1038/s41398-020-01074-z
Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H et al (1998) Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393:702–705. https://doi.org/10.1038/31508
Iida MA, Farrell K, Walker JM, Richardson TE, Marx G, Bryce CH et al (2021) Predictors of cognitive impairment in primary age-related tauopathy: an autopsy study. bioRxiv. https://doi.org/10.1101/2021.06.08.447553
Ikeda K, Akiyama H, Arai T, Oda T, Kato M, Iseki E et al (1999) Clinical aspects of “senile dementia of the tangle type” - A subset of dementia in the senium separable from late-onset Alzheimer’s disease. Dement Geriatr Cogn 10:6–11. https://doi.org/10.1159/000017091
Ikeda K, Akiyama H, Arai T, Sahara N, Mori H, Usami M et al (1997) A subset of senile dementia with high incidence of the apolipoprotein E epsilon2 allele. Ann Neurol 41:693–695. https://doi.org/10.1002/ana.410410522
Jabbari E, Koga S, Valentino RR, Reynolds RH, Ferrari R, Tan MMX et al (2021) Genetic determinants of survival in progressive supranuclear palsy: a genome-wide association study. Lancet Neurol 20:107–116. https://doi.org/10.1016/S1474-4422(20)30394-X
Janocko NJ, Brodersen KA, Soto-Ortolaza AI, Ross OA, Liesinger AM, Duara R et al (2012) Neuropathologically defined subtypes of Alzheimer’s disease differ significantly from neurofibrillary tangle-predominant dementia. Acta Neuropathol 124:681–692. https://doi.org/10.1007/s00401-012-1044-y
Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S et al (2019) Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet 51:404–413. https://doi.org/10.1038/s41588-018-0311-9
Jellinger KA, Attems J (2007) Neurofibrillary tangle-predominant dementia: comparison with classical Alzheimer disease. Acta Neuropathol 113:107–117. https://doi.org/10.1007/s00401-006-0156-7
Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC et al (2016) A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry 21:108–117. https://doi.org/10.1038/mp.2015.23
Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK et al (2017) A unique microglia type associated with restricting development of alzheimer’s disease. Cell 169(1276–1290):e1217. https://doi.org/10.1016/j.cell.2017.05.018
Knopman DS, Parisi JE, Salviati A, Floriach-Robert M, Boeve BF, Ivnik RJ et al (2003) Neuropathology of cognitively normal elderly. J Neuropath Exp Neur 62:1087–1095. https://doi.org/10.1093/jnen/62.11.1087
Kouri N, Ross OA, Dombroski B, Younkin CS, Serie DJ, Soto-Ortolaza A et al (2015) Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun. https://doi.org/10.1038/ncomms8247
Kovacs GG (2015) Invited review: neuropathology of tauopathies: principles and practice. Neuropathol Appl Neurobiol 41:3–23. https://doi.org/10.1111/nan.12208
Kovacs GG, Ferrer I, Grinberg LT, Alafuzoff I, Attems J, Budka H et al (2016) Aging-related tau astrogliopathy (ARTAG): harmonized evaluation strategy. Acta Neuropathol 131:87–102. https://doi.org/10.1007/s00401-015-1509-x
Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC et al (2019) Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 51:414–430. https://doi.org/10.1038/s41588-019-0358-2
Lafirdeen ASM, Cognat E, Sabia S, Hourregue C, Lilamand M, Dugravot A et al (2019) Biomarker profiles of Alzheimer’s disease and dynamic of the association between cerebrospinal fluid levels of beta-amyloid peptide and tau. PLoS ONE 14:e0217026. https://doi.org/10.1371/journal.pone.0217026
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45:1452–1458. https://doi.org/10.1038/ng.2802
Larsson M, Duffy DL, Zhu G, Liu JZ, Macgregor S, McRae AF et al (2011) GWAS findings for human iris patterns: associations with variants in genes that influence normal neuronal pattern development. Am J Hum Genet 89:334–343. https://doi.org/10.1016/j.ajhg.2011.07.011
Lee SH, Meilandt WJ, Xie L, Gandham VD, Ngu H, Barck KH et al (2021) Trem2 restrains the enhancement of tau accumulation and neurodegeneration by beta-amyloid pathology. Neuron 109(1283–1301):e1286. https://doi.org/10.1016/j.neuron.2021.02.010
Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W (1999) The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Gene Dev 13:1822–1833. https://doi.org/10.1101/gad.13.14.1822
Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD et al (2018) GWAS on family history of Alzheimer’s disease. Transl Psychiatry 8:99. https://doi.org/10.1038/s41398-018-0150-6
Martin L, Latypova X, Wilson CM, Magnaudeix A, Perrin ML, Yardin C et al (2013) Tau protein kinases: involvement in Alzheimer’s disease. Ageing Res Rev 12:289–309. https://doi.org/10.1016/j.arr.2012.06.003
McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A et al (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48:1279–1283. https://doi.org/10.1038/ng.3643
McMillan CT, Lee EB, Jefferson-George K, Naj A, Van Deerlin VM, Trojanowski JQ et al (2018) Alzheimer’s genetic risk is reduced in primary age-related tauopathy: a potential model of resistance? Ann Clin Transl Neur 5:927–934. https://doi.org/10.1002/acn3.581
Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM et al (1991) The consortium to establish a registry for Alzheimer’s disease (CERAD). Part II standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41:479–486
Mitchell TW, Mufson EJ, Schneider JA, Cochran EJ, Nissanov J, Han LY et al (2002) Parahippocampal tau pathology in healthy aging, mild cognitive impairment, and early Alzheimer’s disease. Ann Neurol 51:182–189. https://doi.org/10.1002/ana.10086
Morris JC (1993) The clinical dementia rating (CDR): current version and scoring rules. Neurology 43:2412–2414
Mungas D, Tractenberg R, Schneider JA, Crane PK, Bennett DA (2014) A 2-process model for neuropathology of Alzheimer’s disease. Neurobiol Aging 35:301–308. https://doi.org/10.1016/j.neurobiolaging.2013.08.007
Myers AJ, Kaleem M, Marlowe L, Pittman AM, Lees AJ, Fung HC et al (2005) The H1c haplotype at the MAPT locus is associated with Alzheimer’s disease. Hum Mol Genet 14:2399–2404. https://doi.org/10.1093/hmg/ddi241
Nagy Z, VatterBittner B, Braak H, Braak E, Yilmazer DM, Schultz C et al (1997) Staging of Alzheimer-type pathology: an interrater-intrarater study. Dement Geriatr Cogn 8:248–251. https://doi.org/10.1159/000106639
Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D et al (2019) Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol 18:1091–1102. https://doi.org/10.1016/S1474-4422(19)30320-5
Nelson PT, Abner EL, Schmitt FA, Kryscio RJ, Jicha GA, Santacruz K et al (2009) Brains with medial temporal lobe neurofibrillary tangles but no neuritic amyloid plaques are a diagnostic dilemma but may have pathogenetic aspects distinct from Alzheimer disease. J Neuropathol Exp Neurol 68:774–784. https://doi.org/10.1097/NEN.0b013e3181aacbe9
Nelson PT, Alafuzoff I, Bigio EH, Bouras C, Braak H, Cairns NJ et al (2012) Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J Neuropathol Exp Neurol 71:362–381. https://doi.org/10.1097/NEN.0b013e31825018f7
Nelson PT, Jicha GA, Schmitt FA, Liu H, Davis DG, Mendiondo MS et al (2007) Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J Neuropathol Exp Neurol 66:1136–1146. https://doi.org/10.1097/nen.0b013e31815c5efb
Novak M, Jakes R, Edwards PC, Milstein C, Wischik CM (1991) Difference between the tau-protein of alzheimer paired helical filament core and normal tau revealed by epitope analysis of monoclonal antibodies-423 and antibodies-7.51. P Natl Acad Sci USA 88:5837–5841. https://doi.org/10.1073/pnas.88.13.5837
Otero-Garcia M, Xue Y-Q, Shakouri T, Deng Y, Morabito S, Allison T et al (2020) Single-soma transcriptomics of tangle-bearing neurons in Alzheimer’s disease reveals the signatures of tau-associated synaptic dysfunction. bioRxiv. https://doi.org/10.1101/2020.05.11.088591
Panagiotou OA, Ioannidis JPA, Project G-WS (2012) What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol 41:273–286. https://doi.org/10.1093/ije/dyr178
Panchenko MV (2016) Structure, function and regulation of jade family PHD finger 1 (JADE1). Gene 589:1–11. https://doi.org/10.1016/j.gene.2016.05.002
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909. https://doi.org/10.1038/ng1847
Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP et al (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26:2336–2337. https://doi.org/10.1093/bioinformatics/btq419
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Reiman EM, Arboleda-Velasquez JF, Quiroz YT, Huentelman MJ, Beach TG, Caselli RJ et al (2020) Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat Commun. https://doi.org/10.1038/s41467-019-14279-8
Robinson JL, Geser F, Corrada MM, Berlau DJ, Arnold SE, Lee VM et al (2011) Neocortical and hippocampal amyloid-beta and tau measures associate with dementia in the oldest-old. Brain 134:3708–3715. https://doi.org/10.1093/brain/awr308
Sanchez-Contreras MY, Kouri N, Cook CN, Serie DJ, Heckman MG, Finch NA et al (2018) Replication of progressive supranuclear palsy genome-wide association study identifies SLCO1A2 and DUSP10 as new susceptibility loci. Mol Neurodegener. https://doi.org/10.1186/s13024-018-0267-3
Sanchez-Juan P, Moreno S, de Rojaso I, Hernandez I, Valero S, Alegret M et al (2019) The MAPT H1 haplotype is a risk factor for Alzheimer’s disease in APOE epsilon 4 non-carriers. Front Aging Neurosci. https://doi.org/10.3389/fnagi.2019.00327
Santa-Maria I, Haggiagi A, Liu XM, Wasserscheid J, Nelson PT, Dewar K et al (2012) The MAPT H1 haplotype is associated with tangle-predominant dementia. Acta Neuropathol 124:693–704. https://doi.org/10.1007/s00401-012-1017-1
Scheres SH, Zhang W, Falcon B, Goedert M (2020) Cryo-EM structures of tau filaments. Curr Opin Struct Biol 64:17–25. https://doi.org/10.1016/j.sbi.2020.05.011
Schneider JA, Aggarwal NT, Barnes L, Boyle P, Bennett DA (2009) The neuropathology of older persons with and without dementia from community versus clinic cohorts. J Alzheimers Dis 18:691–701. https://doi.org/10.3233/Jad-2009-1227
Sealey MA, Vourkou E, Cowan CM, Bossing T, Quraishe S, Grammenoudi S et al (2017) Distinct phenotypes of three-repeat and four-repeat human tau in a transgenic model of tauopathy. Neurobiol Dis 105:74–83. https://doi.org/10.1016/j.nbd.2017.05.003
Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao LZ, Luo WJ et al (2017) ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549:523–524. https://doi.org/10.1038/nature24016
Sieberts SK, Perumal TM, Carrasquillo MM, Allen M, Reddy JS, Hoffman GE et al (2020) Large eQTL meta-analysis reveals differing patterns between cerebral cortical and cerebellar brain regions. Scient Data. https://doi.org/10.1038/s41597-020-00642-8
Silva MC, Ferguson FM, Cai Q, Donovan KA, Nandi G, Patnaik D et al (2019) Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. Elife. https://doi.org/10.7554/eLife.45457
Siriwardana NS, Meyer R, Havasi A, Dominguez I, Panchenko MV (2014) Cell cycle-dependent chromatin shuttling of HBO1-JADE1 histone acetyl transferase (HAT) complex. Cell Cycle 13:1885–1901. https://doi.org/10.4161/cc.28759
Smith HL, Mallucci GR (2016) The unfolded protein response: mechanisms and therapy of neurodegeneration. Brain 139:2113–2121. https://doi.org/10.1093/brain/aww101
Steinberg KM, Antonacci F, Sudmant PH, Kidd JM, Campbell CD, Vives L et al (2012) Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet 44:872–873. https://doi.org/10.1038/ng.2335
Strickland SL, Reddy JS, Allen M, N’songo A, Burgess JD, Corda MM et al (2020) MAPT haplotype-stratified GWAS reveals differential association for AD risk variants. Alzheimers Dement 16:983–1002. https://doi.org/10.1002/alz.12099
Stutzbach LD, Xie SX, Naj AC, Albin R, Gilman S, Lee VMY et al (2013) The unfolded protein response is activated in disease-affected brain regions in progressive supranuclear palsy and Alzheimer’s disease. Acta Neuropathol Com. https://doi.org/10.1186/2051-5960-1-31
Van Cauwenberghe C, Van Broeckhoven C, Sleegers K (2016) The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet Med 18:421–430. https://doi.org/10.1038/gim.2015.117
Voas MG, Rebay I (2003) The novel plant homeodomain protein rhinoceros antagonizes Ras signaling in the Drosophila eye. Genetics 165:1993–2006
Walker JM, Richardson TE, Farrell K, Iida MA, Foong C, Shang P et al (2021) Early selective vulnerability of the CA2 hippocampal subfield in primary age-related tauopathy. J Neuropath Exp Neur 80:102–111. https://doi.org/10.1093/jnen/nlaa153
Wesseling H, Mair W, Kumar M, Schlaffner CN, Tang S, Beerepoot P et al (2020) Tau PTM profiles identify patient heterogeneity and stages of Alzheimer’s disease. Cell 183(1699–1713):e1613. https://doi.org/10.1016/j.cell.2020.10.029
Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26:2190–2191. https://doi.org/10.1093/bioinformatics/btq340
Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M et al (2001) Tauopathy in drosophila: neurodegeneration without neurofibrillary tangles. Science 293:711–714. https://doi.org/10.1126/science.1062382
Yamada M (2003) Senile dementia of the neurofibrillary tangle type (tangle-only dementia): neuropathological criteria and clinical guidelines for diagnosis. Neuropathology 23:311–317. https://doi.org/10.1046/j.1440-1789.2003.00522.x
Yokoyama JS, Karch CM, Fan CC, Bonham LW, Kouri N, Ross OA et al (2017) Shared genetic risk between corticobasal degeneration, progressive supranuclear palsy, and frontotemporal dementia. Acta Neuropathol 133:825–837. https://doi.org/10.1007/s00401-017-1693-y
Yu L, Chibnik LB, Srivastava GP, Pochet N, Yang J, Xu J et al (2015) Association of Brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol 72:15–24. https://doi.org/10.1001/jamaneurol.2014.3049
Zeng LL, Bai M, Mittal AK, El-Jouni W, Zhou J, Cohen DM et al (2013) Candidate tumor suppressor and pVHL partner Jade-1 binds and inhibits AKT in renal cell Carcinoma. Cancer Res 73:5371–5380. https://doi.org/10.1158/0008-5472.Can-12-4707
Zhou MI, Foy RL, Chitalia VC, Zhao J, Panchenko MV, Wang HM et al (2005) Jade-1, a candidate renal tumor suppressor that promotes apoptosis. P Natl Acad Sci USA 102:11035–11040. https://doi.org/10.1073/pnas.0500757102
Zhou MI, Wang H, Ross JJ, Kuzmin I, Xu C, Cohen HT (2002) The von Hippel-Lindau tumor suppressor stabilizes novel plant homeodomain protein Jade-1. J Biol Chem 277:39887–39898. https://doi.org/10.1074/jbc.M205040200
Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR et al (2020) Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat Med 26:131–142. https://doi.org/10.1038/s41591-019-0695-9
Acknowledgements
The authors would like to acknowledge the neuropathology core of the Massachusetts Alzheimer Disease Research Center, the Biosample Management Repository at Genentech/Roche, the brain repository at UCI, Knight Alzheimer Disease Research Center Neuropathology Core at Washington University School of Medicine, the BioRepository and Integrated Neuropathology Laboratory and Precision Neuropathology Core at the University of Washington, the neurodegenerative disease brain bank at the University of California San Francisco, the Neuropathology Brain Bank & Research Core at Mount Sinai, the Newcastle Brain Tissue Resource, and the following people: Ryan Cassidy Bohannan, Chad Caraway, Allison Beller, Kim Howard, Suresh Selvaraj, Ward Ortmann, Ping Shang, Jeff Harris, and Chan Foong. The results published here are in whole or in part based on data obtained from the AD Knowledge Portal (https://adknowledgeportal.org).
Funding
IMSSM: R01 AG054008, R01 NS095252, R01 AG060961, and R01 NS086736 Rainwater Charitable Foundation, Genentech/Roche, Alexander Saint-Amand Fellowship (JFC), F32 AG056098 and P30 AG066514 (KF), P50 AG005138 and P30 AG066514 (VH, JFC, MS, SG, AG, PRH), 75N95019C00049 (VH), K99 AG070109 (SJA), U01 AG058635 (AG, EM, AER) BU / MSSM / MAYO: R01 AG062348 (AM JFC DD), UMC: P50AG008702 (JPV AFT), BU: U54 NS115266 (AM), R01 CA079830 (HTC), UPENN: P30 AG010124, P01 AG017586 and U19 AG062418 (JQT), P30 AG072979 and P01 AG066597 (EBL), R01 AG066152 (CTM), PITT: R01 AG066152 P30 AG066468 (JK), Banner: U24 NS072026 and P30 AG019610 The Arizona Department of Health Services, and the Michael J. Fox Foundation for Parkinson’s Research (TB), Johns Hopkins: P50 AG05146 (JCT), U Iowa: K23 NS109284, The Roy J. Carver Foundation, the Carver College of Medicine, and the Williams-Cannon Foundation (MMH), Northwestern: P30 AG013854 (MEF), Emory: P30 NS055077 and P50 AG025688 (MG), OHSU: P30 AG08017 (RW), UTSW: Winspear Family Center for Research on the Neuropathology of Alzheimer Disease (CWIII), Toronto: Rossy Foundation and by the Safra Foundation (GGK), Stanford: R01AG059848 (IC), MADRC: P50 AG05134, P30 AG062421 (BTH), RUSH: ADC grant AG10161 and MAP grant (JS), UCI: R01AG021055 and P50AG016573 (CHK, MMC), P01AG000538 (WP), UCSD: P30 AG062429 01 and P50 AG005131 (RR), UK: P30 AG028383 (PN), U Washington: P50 AG005136, P30 AG066509, U01 AG006781, U19 066567 and the Nancy and Buster Alvord Endowment (CDK), Washington U / Knight ADRC: P30 AG066444, P01 AG003991, 01 AG026276 (RJP), Newcastle: UK Medical Research Council (G0400074), by Brains for Dementia research, a joint venture between Alzheimer’s Society and Alzheimer’s Research UK and by the NIHR Newcastle Biomedical Research Centre awarded to the Newcastle upon Tyne Hospitals NHS Foundation Trust and Newcastle University (JA), Other: J.M.R. Barker Foundation, The McCune Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
JFC, DWD, MF, PRH, GGK, EBL, PTN, JQT, are editorial board members and JA is editor in chief of Acta Neuropathologica, but were not involved in the editorial handling of this article. JH, TB, are employees of Genentech (a subsidiary of Roche) and hold stocks/stock options in FH-LR Ltd. AG is on the Scientific advisory board for Genentech and consultant for AbbVie. All other authors declare no relevant conflicts.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material. All code is available by request of the corresponding author
Rights and permissions
About this article
Cite this article
Farrell, K., Kim, S., Han, N. et al. Genome-wide association study and functional validation implicates JADE1 in tauopathy. Acta Neuropathol 143, 33–53 (2022). https://doi.org/10.1007/s00401-021-02379-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-021-02379-z